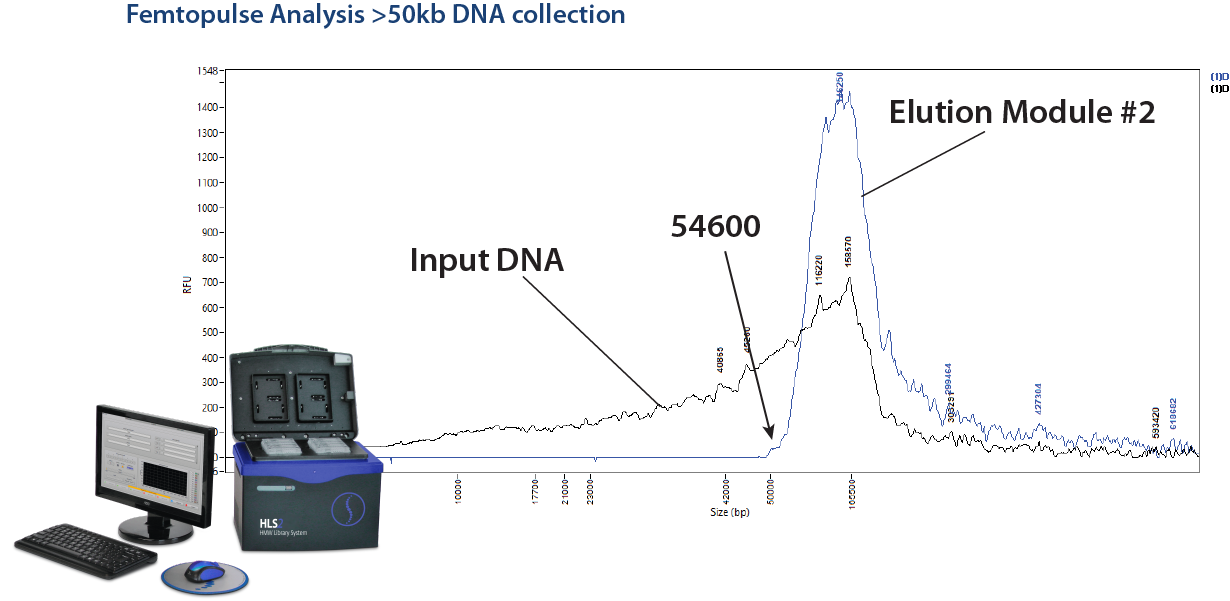
We have been getting quite a few inquiries of late from researchers who would like to size select DNA fragments above 50kb. This may be due in no small part to our friends at Grandomics, China, who have been citing this capability in a number of interesting collaborative studies (see the reference links below). In short, this is something that cannot be accomplished on a BluePippin, and Grandomics has been using the HLS2 platform to accomplish this feat. Here at Sage, we have compiled a cassette kit (HSS-0012 or HSS-0004) specifically for this, and for DNA size selection in general. In fact, with direct current electrophoresis, the HL2 can size select DNA in ranges between 1-20kb, and with pulsed-field protocols it can size select Ultra-HMW DNA into the hundreds of KB and up to 2MB.
HLS2 Size selection on the HLS2 is outlined in this application note: Ultra-HMW DNA Size Selection with the HLS2 instrument and a >50kb Hi-Pass protocol. A quick note to Oxford Nanopore users: this method uses a starting input of 5 ug of DNA and yields about 1-1.5 ug of DNA, while some Oxford Nanopore input requirements may be quite a bit higher.
References:
Huang, Z., et al. 2023. Evolutionary analysis of a complete chicken genome. PNAS 12 (8) e2216641120.
https://doi.org/10.1073/pnas.2216641120
Morita, S., et al. 2023. The draft genome sequence of the Japanese rhinoceros beetle Trypoxylus dichotomus septentrion towards an understanding of horn formation Sci Rep 13, 8735 (2023).
https://doi.org/10.1038/s41598-023-35246-w
Xing Guo, et al. 2023. The genome of Acorus deciphers insights into early monocot evolution. Nat Commun 14, 3662 (2023).
https://doi.org/10.1038/s41467-023-38836-4
Kun Li, et al. 2023. Genetic Diagnosis of Facioscapulohumeral Muscular Dystrophy Type 1 Using Rare Variant Linkage
Analysis and Long Read Genome Sequencing. medRxiv preprint.
https://doi.org/10.1101/2023.06.05.23290975
Q Wang, et. al. 2023. Draft genome of the oriental garden lizard (Calotes versicolor). Front. Genet., 20 February 2023
Sec. Livestock Genomics Volume 14 – 2023
https://doi.org/10.3389/fgene.2023.1091544
Fengjiao Ma, et al. 2023. Gap-free genome assembly of anadromous Coilia nasus. Sci Data 10, 360 (2023).
https://doi.org/10.1038/s41597-023-02278-w