Range+T Size Selection
Note: Range + T is available for all PippinHT instruments. However, only BluePippins with serial numbers 2700 and above will run the method.
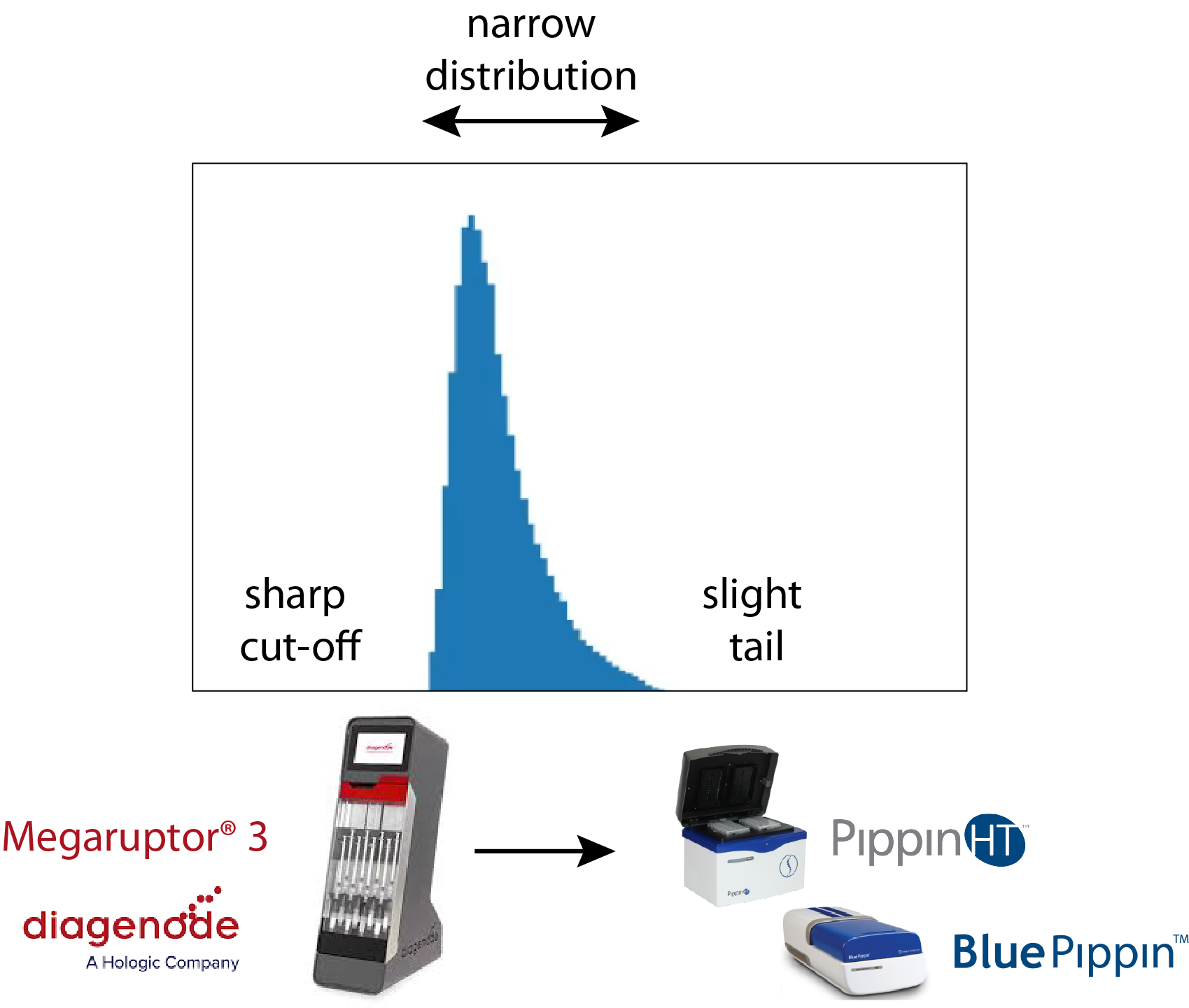
Sage’s DNA size selection technology has been used to help improve PacBio sequencing, particularly for mammalian and complex genome work since the introduction of the BluePippin (2012). Over the last few years, PacBio®’s Hi-Fi circular consensus sequencing has been the method that has set the gold standard for whole genome sequencing. In terms of library size, this technique benefits greatly from a sharp size cut-off at the LMW end of the size distribution (~10kb) and a slight tail at the HMW end. It also benefits from a narrow fragment size distribution around 17-18kb. A common way to accomplish this is to shear DNA with Diagenode’s Megaruptor®3 device, followed by size selection on a PippinHT or BluePippin.
To get the desired fragment distribution at these size ranges is not trivial. In Sage gel cassettes, the total input load – and the tight fragment distribution profile of Megaruptor 3 sheared samples – affects the mobility of the DNA in a manner that cannot be easily accounted for with our standard “Range” programming method. Essentially, the start of collection of DNA is accurate and straightforward, while the timing for the end of collection can be affected by the physical nature of the sheared DNA sample. To this end, we developed a new size selection method called “Range+T”. With this method, the base pair start of collection is entered in the software and the end of collection is determined by entering a time for elution. Through a little trial and error, a time setting can be determined to overcome calibration uncertainty for the end of collection. We conducted an early access program with the Copetti lab at the Arizona Genomics Institute, resulting in increased PacBio HiFi read lengths, that you can read about in this application note.
A Collaboration with Diagenode
We decided to do a deep dive into Range+T to get a better handle the method, and to develop best practices for using Diagenode Megaruptor 3 and PippinHT or BluePippin. Working with Diagenode, we were able to process many DNA samples and experiment with multiple settings. We outlined our findings in a new application note, Best practices: Sage Science size selection with Diagenode Megaruptor shearing for long-read sequencing library preparation . We encourage you to take a look if you are interested in working with these size ranges. We found that careful coordination of Megaruptor shearing and Sage size-selection conditions are critical:
1. Megaruptor shearing conditions should ideally be chosen to provide a sheared DNA input with an average size at, or slightly above, the desired library size.
2. For accurate LMW cut-offs, start of collection DNA size on PippinHT or BluePippin should be less than or equal to the peak size (mode) of the sheared DNA input.
3. The upper boundary of the size distribution will be determined by the Megaruptor shear conditions if long Sage Range+T elution times are used (i.e., 30 minutes or longer).
4. Sage Range+T programming can provide significantly narrower size distributions by shortening the Range+T elution time below 30 minutes.
For size selection, provided the same Megarupter shear conditions (and input amount) are used, we recommend starting with Time values at 5 minute intervals (ie. 5,10,15,20 minutes) to determine the best distribution conditions. It is very important to enter a starting base pair value that is before the mode of the Megaruptor distribution – so QC your samples!
Range+T is available now for both PippinHT and BluePippin. It requires a software upgrade (v.6.41 and v1.14, respectively, available for download on sagescience/support). We have also packaged cassette kits specifically for this purpose; HRT7510 and BRT7510. Starting Range values can be between 9-30kb.