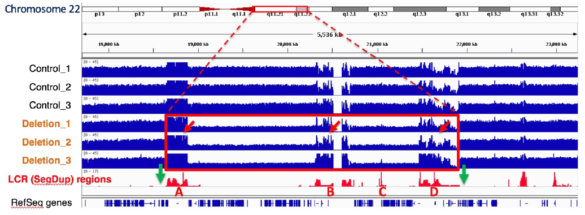
An international collaboration led by the Stanford labs of Alexander Urban and Hanlee Ji, used the SageHLS system to map a Mb-sized deletion in a complex region of the human genome, 22q11.2, that is associated with a variety of neurodevelopmental and neuropsychiatric disorders. Structural variation (SV) in this region is difficult to study because the breakpoints are frequently located in four large segmental duplications (up to 400kb in length) that are scattered over a 3 Mb area. Conventional short-read sequencing cannot unambiguously identify the breakpoints of these SVs because of the high homology between the segmental duplications. The researchers designed an HLS-CATCH procedure in which Cas9 was used to cleave unique sites outside of the 3 Mb region that contains the segmental duplications. In a patient sample carrying a large deletion, a ~300kb CATCH product from the deletion allele could be cleanly separated from the much larger 3 Mb CATCH product from the wild-type allele by preparative pulsed-field electrophoresis in the SageHLS. The researchers were then able to sequence the 300kb deletion product using Oxford Nanopore long-read methods and successfully map the deletion breakpoints.
Some of this work was presented in a scientific poster at the recent AGBT 2020 conference at Marco Island. The poster can be viewed here.
The green arrows below indicate the Cas9/RNA target sites that flank the 22q11.2 region (Bo Zhou, et al. presented at AGBT 2020)