A new preprint has landed on BioRxiv that reports on high-accuracy circular consensus sequencing (CCS) on the PacBio Sequel. The study, “Highly-accurate long-read sequencing improves variant detection and assembly of a human genome”, is authored by PacBio and an impressive team of collaborators featuring notable bioinformaticians and members of the Genome in a Bottle Consortium. The data suggest that with CCS, very accurate (Q30) DNA sequence can be obtained from a single >10kb molecule (read PacBio’s blog on the study here)
The gist of the method is this: processivity improvements have yielded polymerase read lengths of approximately 150kb. Since SMRTbells are circular, a polymerase should be able pass a 15kb DNA fragment 10 times, and re-reading the molecule 10 times should yield a 99.9% accurate sequence.
 Our customers may be aware of our High-Pass library size selection with the BluePippin in which, average read lengths can be improved – often doubling N50s. But for CCS, its crucial to prepare libraries with relatively uniform size, with the goal of producing a full run of Q30 15kb reads to be assembled against a reference. For this, the SageELF DNA fractionator is the tool for the task. The SageELF produces narrow fragment size distribution, reproducibly, and provides a great deal of flexibility. For instance, users can run a 15kb library and archive a 20kb library from another well. Or, adjacent wells can be pooled, increasing library amount with only a slight widening of the distribution range.
Our customers may be aware of our High-Pass library size selection with the BluePippin in which, average read lengths can be improved – often doubling N50s. But for CCS, its crucial to prepare libraries with relatively uniform size, with the goal of producing a full run of Q30 15kb reads to be assembled against a reference. For this, the SageELF DNA fractionator is the tool for the task. The SageELF produces narrow fragment size distribution, reproducibly, and provides a great deal of flexibility. For instance, users can run a 15kb library and archive a 20kb library from another well. Or, adjacent wells can be pooled, increasing library amount with only a slight widening of the distribution range.
In BioRXiv paper, the follow protocol was used:
1. Start with 3-4 ug DNA
2. Shear the DNA with Diagenode Megarupter to 15-20kb
3. Construct SMRTbell libraries
4. Size Fractionate with the SageELF, collect a 15kb fraction
5. Run on the Sequel
The following fractionation protocol was used (this is not detailed in the publication, but based on private communication with the authors):
1. Use cassette kit #ELD7510 (for 1-18 kb fractionation)
2. Load 1-2 ug/run (fractions were pooled from two runs)
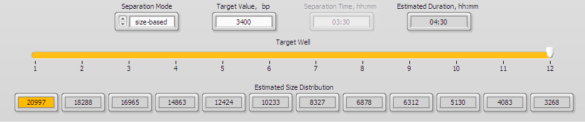
3. Enter 3400 into well 12 in the protocol editor (below)
4. A 15 kb fraction will be found in well 4, approx. 4.5 hour run
We do have alternative recommendations:
1. We offer cassette kit #ELD4010 (for 10-40kb fractionation). This should provide an even narrower size distribution than the 0.75% agarose cassettes.
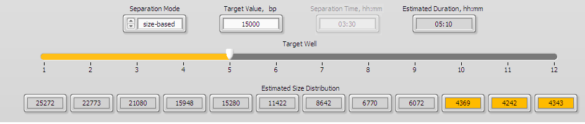
2. Enter 15000 into well 5 in the protocol editor. The 15 kb fraction will be found in well 5 (and a 20kb fraction will be found in well 3).
3. 3-4ug of input DNA in one lane should be sufficient so pooling from two runs may not be needed.