We here at Sage Science are excited to have joined forces with Universal Sequencing Technology to promote their TELL-Seq barcode linked-read technology. One of the reasons for our excitement is that TELL-Seq, and the transposase-based barcoding method benefits greatly from high molecular weight DNA – one of Sage’s areas of expertise. Our own HLS-CATCH method is one area that is a particularly nice fit with TELL-Seq particularly given the relatively small amount of DNA that HMW target purification yields. The easy library workflow and ready access to Illumina sequencing is also a big plus.
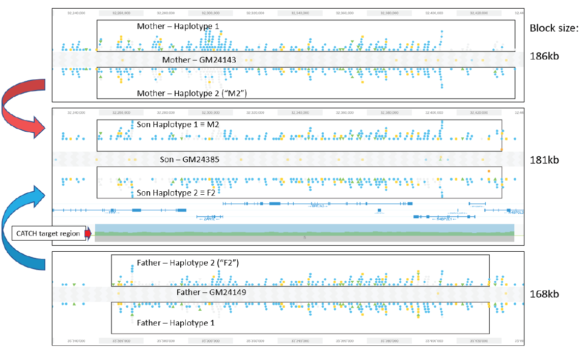
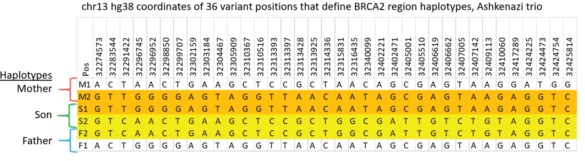
Validating this proposition was simple and straightforward. We performed HLS-CATCH using cultured cells from the Genome in a Bottle Ashkenazi trio sample set using a Cas9 guides that we designed to purify a 187kb target containing the BRCA2 locus. TELL-seq libraries were prepared from the targets from each individual, sequenced and run through 10X Genomic’s Long Ranger analysis package and then visualized the data on the 10X Loupe browser. The phase blocks sizes were 186 kb (mother), 181kb ( son), and 168Kb ( father), and only a small-genome scale analysis was required. Download our whitepaper here.
The variants in the 6 haplotypes compared favorably to the Genome in a Bottle high quality sequences.
Clearly CATCH and TELL-seq can be an economical alternative for obtaining long range genomic data for gene targets. The value of this technique was previously demonstrated in a Stanford University paper (Shin, GW, et al. 2019) where, among other analyses, the 4Mb MHC histocompatibility complex was phased using the 10X Genomics linked read method. The simplicity of TELL-Seq is great news vis a vis the effort required for long-read sequencing libraries. However, strides are being made there as well. Two recent papers have demonstrated great results using HLS-CATCH with PacBio sequencing (Walsh, T. et al. 2020) and Oxford Nanopore (Zhou, B, et al. 2020).