Recently we blogged about the rise of cell-free DNA studies, and how precise size selection is one tool that can help researchers isolate DNA of interest for further analysis (such as sorting out fetal from maternal genetic material).
At the AGBT Precision Health meeting in Scottsdale, Ariz., last month, we teamed up with Rubicon Genomics to present a poster demonstrating how our technologies can be used together to generate better results from cell-free DNA studies. (If you were at the meeting, it was poster #107.)
For this work, we performed pre-library size selection using the Pippin Prep to separate fragments 160 bp and larger (since that’s about the size of the mononucleosomal unit) from smaller fragments, which are typically the ones scientists want to analyze for cancer or prenatal studies. After sizing, we prepared libraries for sequencing using Rubicon Genomics’ ThruPLEX® Plasma-Seq kit, which is designed for low-input samples from plasma and liquid biopsies like cfDNA. Libraries were also prepared and analyzed without size selection so we could see how much of a difference that step made.
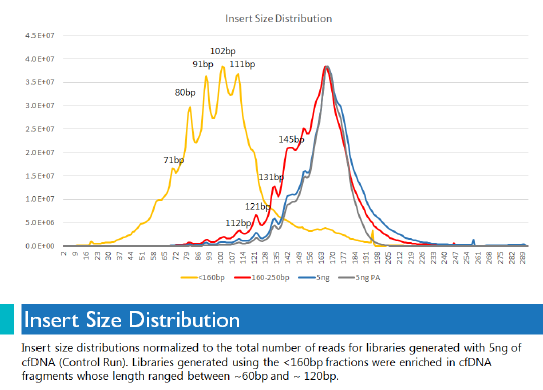
Even though we started with less than 1 ng of size-selected material, we were able to produce highly diverse libraries that were significantly enriched for cell-free DNA fragments between 60 bp and 120 bp. That enrichment did not happen for libraries prepared without size selection. This graph of insert size distribution shows how clearly delineated the shorter fragments were when size selection was used.
While Pippin Prep was used for this project, Sage customers can use PippinHT as well to perform what we call “low-pass” sizing — that is, removing everything larger than a certain size. We look forward to seeing how our users deploy automated size selection technology to enhance their cell-free DNA studies.





