Well, we have another fabulous AGBT behind us that we were happy to attend. Our take home messages? A. Spatial transcriptomics is the exciting new frontier. B. True human WGS has been accomplished and is moving into population-scale work. And C. There are a lot of new sequencing technologies being bandied about.
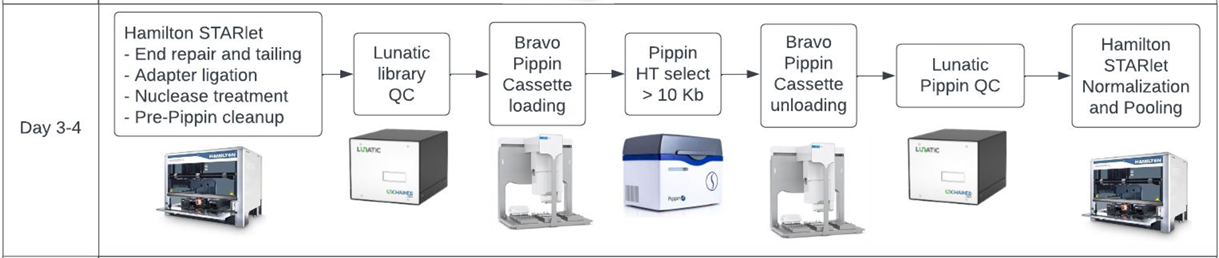
Though we weren’t presenting or introducing, Sage Science did have a few key mentions. The Broad Institute described their efforts to automate a PacBio HiFi whole genomes sequencing pipeline for the NIH’s All of Us program. In their poster, you can view it here, they go into quite a bit of detail why gel size selection (via the PippinHT) is the preferable method with HiFi library construction.
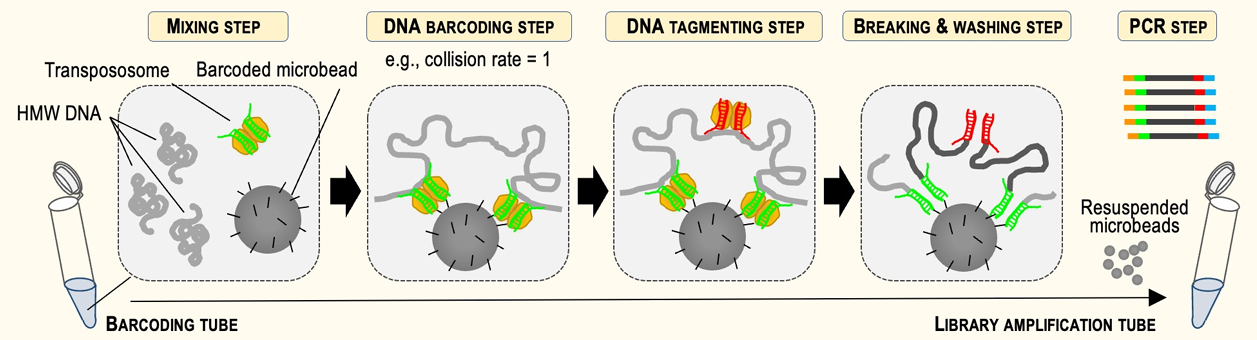
HLS-CATCH, our method for enriching high molecular weight targets made an appearance in a few posters. Universal Sequencing, our frequent collaborators and partners (we also sell their TELL-seq kits) presented a nice demonstration of their linked-read technology. They show haplotype phasing data for larger targets (200kb with HLS-CATCH) and smaller targets (4-20kb) using PCR and andCas9 Exo/Pulldown method (a modified CaBagE protocol).
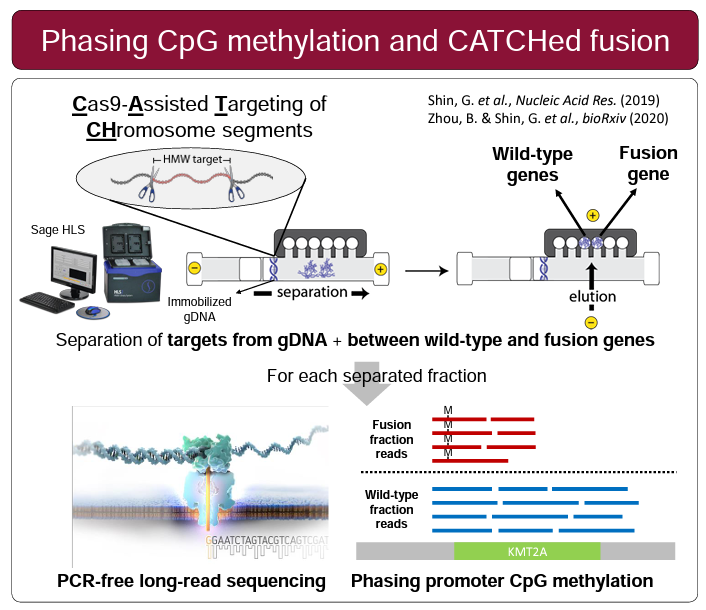
Three posters were presented that featured HLS-CATCH and Oxford Nanopore sequencing. Researchers from the Ji lab at Stanford University presented a very compelling poster on KMT2A translocations, a common chromosomal abnormality in leukemias. Using HLS-CATCH (they use the term CLTR-Seq) and nanopore sequencing they identify rearrangement structure as well as the methylation landscape. Stanford researchers from the Ji and Urban labs presented a poster on a study in which HLS-CATCH is used to sequence segmental duplications, also by nanopore sequencing, associated with the neuropsychiatric 22q11.2 deletion region. Finally, CATCH was used in a study presented by Gavin Arno from the University College London and collaborators that is focused on inherited retinal disease and complex rearrangements associated with the OPN1LW/OPN1MW gene array. The authors, like the Stanford researchers, conclude that CATCH-nanopore sequencing is effective unraveling genomic mysteries that are “intractable to NGS”.