NextOmics/GrandOmics is the largest third-generation sequencing service company in China. The NextOmics division focuses on animal, plant and bacterial genome sequencing, while GrandOmics focuses on human genome sequencing.
GrandOmics became the first certified PromethIon service provider in China in June, 2018. In April, 2019, GrandOmics and Oxford Nanopore announced a strategic collaboration to sequence 100,000 Chinese genomes on the PromethIon platform by the end of 2021.
Recently, NextOmics/GrandOmics has disclosed some examples of its PromethIon output on Twitter (see below), and shared with Sage Science an overview of their whole genome workflow.
1) Input material – human cultured cell lines, young plant leaves.
2) Extraction is carried out using a modified phenol/chloroform method.
3) For ultra-long reads, size selection is performed using the SageHLS instrument.
4) SageHLS input: not more than 10ug per lane.
5) SageHLS size selection program(options, depending on extraction quality): High-Pass 50kb, 100kb, 250kb, 300kb, 350kb, 500kb etc. (collection stage only)
6) Oxford library kit: SQK-LSK109 Ligation kit.
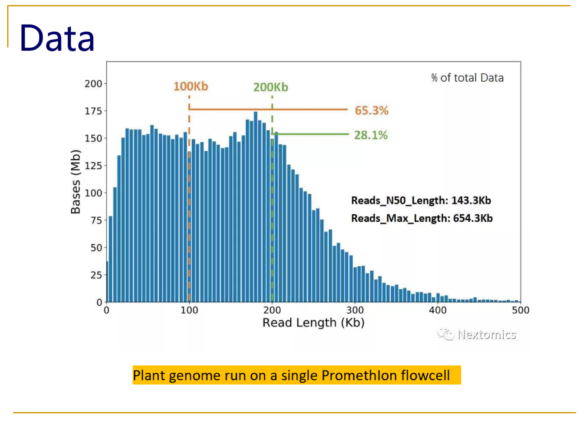
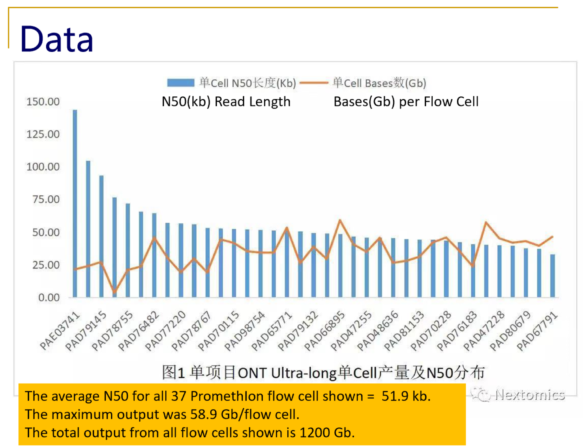
Typical size distributions from the PromethIon flow cell show read N50 values as high as 140kb, with an impressive 28% of the reads longer than 200kb.
[click the images to enlarge]
What NextOmics/GrandOmics says about SageHLS for ultra-long read PromethIon sequencing:
“The bottleneck of ONT sequencing is sample preparation, especially for ultra-long sequencing. The ONT user should optimize the extraction steps in order to protect the DNA from shear forces generated during liquid handling. After the extraction, we use SageHLS size-selection to remove shorter DNA fragments and enrich the ultra-high molecular weight DNA. SageHLS is a key component of our ultra-long PromethIon workflow.”
**We would like to express our gratitude to our great distributor, APG Bio, for their help with this post!