We’ve just released new dye-free cassettes for the BluePippin and Pippin Prep that use dye-labeled DNA markers* as internal standards, instead of typical external marker set. In the new product, the labeled markers will come premixed wi
th the Ficoll loading solution, and will be mixed and loaded with the sample DNA in each lane. The internal standards are designed to run well ahead of sample DNA targeted for elution, so that little or no marker contamination of eluted samples should occur.
These new cassettes will offer the following benefits:
- Faster run times. Internal standards automatically correct for the lane-to-lane mobility variation that occurs at higher voltage. Using the new high voltage 1.5% cassettes, collection of DNA fractions as large as 500 bp can be carried out in 35 minutes or less.
- Improved accuracy and reproducibility. Compared to dye-free cassettes run with an external marker, the accuracy and reproducibility of DNA extractions** will be improved (~ 5% vs. 10%). We have found the reproducibility to be quite good:
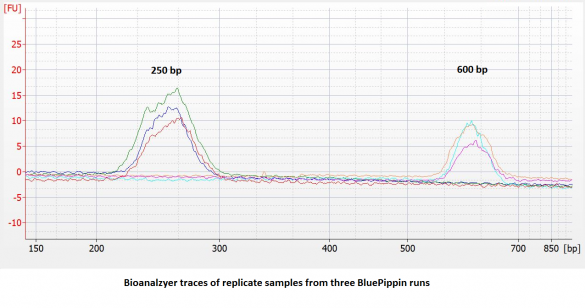
- 25% higher sample capacity. All five lanes can be used for samples. (This also means the per-sample cost goes down by 20%.)
- Use fewer lanes per run without penalty. Users may run a single sample, re-tape the open ports and save the remaining 4 lanes for later. With external standards, each run requires at least 2 lanes: one for external marker and one for sample.
Will internal standards contaminate your sequencing result?
The BluePippin internal standards are designed to run well ahead of the usable size range of the cassette. This minimizes the chance for marker contamination in eluted sample DNA since the platform excels at eliminating sample contamination from lower molecular weight fractions. In addition, when fractionating adaptered libraries, sequencing artifacts caused by marker DNA contamination are expected to be minimal, since the marker fragments are not complementary to amplification primers used by the major sequencing platforms. This expectation has been demonstrated on the Illumina platform (Tech Note in process). To enable identification/filtering of marker sequence, we’ve posted the sequences on our support page (www.sagescience.com/support). You can click the link below to get there:
http://sagescience.com/wp-content/uploads/2014/08/Internal-standard_sequences-UPDATED-082014.pdf
Input load and accuracy
The 1.5% DF cassettes can accommodate up to 10 ug of genomic input DNA. However, there are load-dependent changes in DNA mobility in these cassettes. Since most of our customers are running DNA samples of 2ug or less, we have changed our standard input load for calibration to 2 ug. Most users will find excellent accuracy for input loads up to 2 ug. At higher input loads, users should be prepared to run some preliminary tests to determine the best settings for their application. In general, at loads greater than 2 ug, selected DNA size will be slightly higher than programmed. As input load increases, so will the deviation from the programmed value.
Look or ask for these cassettes for the BluePippin:
For the BluePippin
- BDF1510 – 1.5% agarose with internal standards for the BluePippin. 250 bp-1.5 kb.
- BDF3010 – 3% agarose with internal standards for the BluePippin. 90 bp-300 bp.
For the Pippin Prep
- CDF1510 – 1.5% agarose with internal standards for the Pippin Prep. 250 bp-1.5 kb.
- CDF3010 – 3% agarose with internal standards for the Pippin Prep. 90 bp-300 bp.
* We use Mirus fluorescent label with our markers. “www.mirusbio.com”
** Accuracy = Deviation of the actual target base pair value from software input value divided by the actual value expressed as a percentage. (In calibration, actual DNA sizes are determined using an Agilent Bioanalyzer 2100.)
Reproducibility = 2X standard deviation of replicate samples expressed as a percentage of the average value.






2 Responses to New Cassettes Use Internal Standards