We’re pleased to be shipping our Mid-Range size selection cassette (BMF7510) for the BluePippin. While our Low-Range cassette (BLF7510, 1kb-10kb) works for many mate-pair library construction protocols, we’ve been hearing from our customers that there is interest in creating larger circularized molecules. This cassette, which continues to extend the range and flexibility of the BluePippin product line, should serve as a useful tool for large-fragment preps and help produce more diverse and meaningful data.
(Some internally generated validation data can be found on this pdf.)
In this blog, we’d like to outline some characteristics on the product to help users develop appropriate methods.
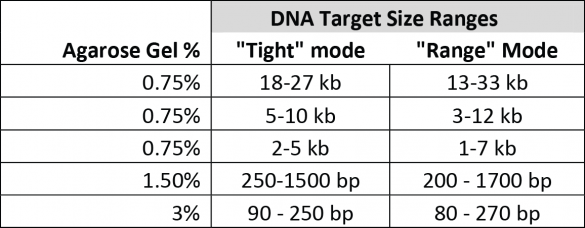
CVs and target ranges
We define all target ranges in terms of what a user can accurately collect by entering a value in “tight” mode using our software. We also have a specification, “Minimum Size Distribution as Expressed by CV,” that describes the minimum distribution of fragments that may be collected given the capabilities of the system and the resolution of the gel. For instance, as a rule of thumb, a CV of 8% (our spec for smaller targets) will yield a range of fragments that are at most + 16% of the median fragment. However, the mid-range targets collect at a comparatively larger distribution minimum (20% CV), which limits the effective range of accurate “tight” cuts. This chart compares the accurately selectable “tight mode” ranges to the “range mode” ranges for the gel cassette calibrations available for the BluePippin at this time:
Sample Load Dependence
To calibrate the mid-range cassettes, we use a 5 ug sample load of sheared E. coli genomic DNA. Higher sample loads (up to 10ug) will usually run somewhat slower, relative to the marker DNA, and for this reason, you will need to program higher bp target or range values in the protocol to collect a desired size fraction. In our tests, when using a 10 ug input load, bp target values should be increased by 10-15% to compensate for the changes in mobility caused by the increased load.
Another factor to consider is the size distribution of the input DNA. For calibration and testing, we use an E. coli genomic DNA sample that has a very broad, almost flat size distribution. Using such samples simplifies our calibration process, which involves accurately sizing fractions sliced from the flat input samples. However, most input samples for mate-pair libraries are generated by methods that produce much narrower size distribution (DigiLab Hydroshear, for instance). Such samples will therefore show a different mobility dependence on input load than our E. coli genomic DNA test samples. For this reason, customers should plan on conducting some pilot experiments on non-valuable samples to investigate the mobility-load relationship produced by their specific library protocol.
Bottom line on 0.75% Mid-range cassette definition
- BluePippin 0.75% mid-range cassettes facilitate size fractionations of DNA samples out to 30kb.
- The CVs of tight size selections will be wider than those obtained from other BluePippin and Pippin Prep cassettes (mid-range CVs around 20%).
- The mid-range protocols show a significant mobility dependence on input load. Sage uses a 5 ug genomic sample with a broad size distribution for calibration. Higher input loads and samples with narrow size distributions will require user optimization of programmed size values for accurate results.
- Input loads higher than 10 ug per lane will have unpredictable results, and are not recommended.
If you plan to use the the BluePippin for mate-pair library construction, we’d love to hear of your progress or suggestions. To share, please contact us at info@sagescience.com or reply to this blog.
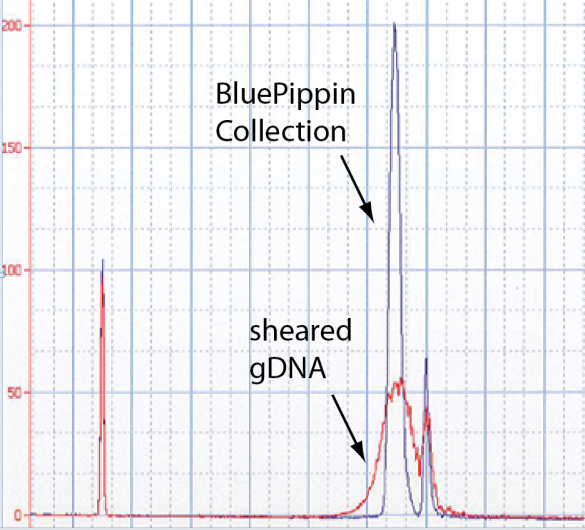
The image below is an Agilent Bioanalyzer trace of a nice 8 kb collection in the middle of a gDNA shear. This is near the limit of the Bioanalyzer’s detection target range.