Back from Baltimore and the Stellar PacBio UGM
Earlier this week Pacific Biosciences hosted a user group meeting at the University of Maryland in Baltimore, and we were pleased to participate in and co-sponsor the event. As our blog readers know, we recently announced a co-marketing partnership with PacBio to provide BluePippin size selection to PacBio users to help them generate longer average read lengths.
At the user group meeting, about 100 attendees came to discuss their own experiences with the sequencing platform and to learn how other people were pushing the boundaries on read length, DNA input requirements, and more. It was great to see so much enthusiasm for PacBio’s sequencer. This is a really engaged community — almost every presentation was followed by several questions and often lively discussion within the audience as people shared their own methods or results.
A key theme at the meeting revolved around the dramatic improvements in throughput and read length since the PacBio system first launched. Speakers got in the habit of noting what year certain data had been generated as shorthand to help attendees understand how the data might look today if the experiment were run with newer enzymes, reagents, or hardware. A speaker from Baylor noted that since receiving their instrument, throughput has increased more than 10-fold while average read length has jumped from 1.5 Kb to 7 Kb. Here at Sage, we are proud to be part of that growth in read length — and we were glad to see speakers reporting a significant boost in average read length with the incorporation of BluePippin into the workflow.
With these extraordinarily long reads, many of the speakers presented information on genetic elements or attributes that have never been seen before, including connections of distant tandem repeats, genome-wide methylation in pathogens, and fully sequenced mRNA transcripts. There was also quite a bit of excitement around moving toward de novo mammalian sequencing with the PacBio workflow, which can currently perform de novo sequencing and automated assembly/finishing of microbial genomes.
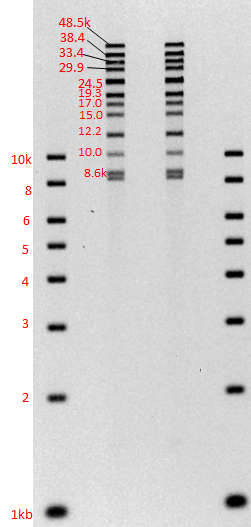
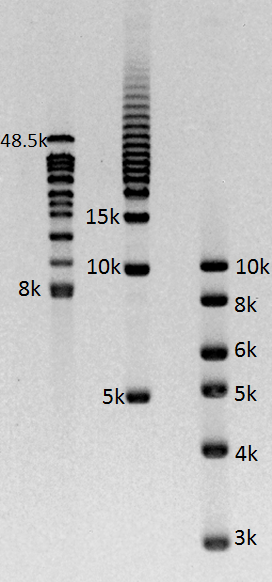
From Sage Science, Chris Boles offered an update on BluePippin for PacBio. (For the current protocol, click here.) He presented data from one genome center showing a project in which the N50 median read length of 4,800 bases was boosted to 9,100 bases by adding BluePippin to remove smaller fragments from the library. Boles also reported that we are working to develop additional protocols with PacBio to keep extending those N50 numbers. We look forward to continuing to help PacBio users generate even more impressive read lengths from their sequencers!
ASMS, Here We Come!
We are packing our bags for Minneapolis and the 61st annual conference of the American Society for Mass Spectrometry. As scientists who come from the DNA realm, the protein world is very exciting to us and we’re looking forward to one of the premier meetings in this field.
It’ll be great to see the award presentation from Dick Smith, head of proteomics research at Pacific Northwest National Laboratory. Dick has a long history of optimizing mass spec technology, so no doubt his talk will cover his greatest hits as well as what’s new and intriguing in his lab today. We’re also interested in hearing about cutting-edge work in top-down proteomics and other approaches during the regular conference sessions.
This is the first year Sage will have a booth at ASMS, so please stop by booth #31 and say hello! We’ll be introducing our two new automated tools for the protein market: the BluePippin for proteins and the Sage ELF. BluePippin, one of our DNA sizing platforms, has new cassettes to enable targeted SDS-protein collection of a desired fraction from up to five samples per run. The ELF, or Electrophoretic Lateral Fractionator, allows users to fractionate a protein sample into 12 contiguous size fractions. These tools provide increased reproducibility and ease-of-use for procedures that require gel isolation of proteins, including top-down and bottom-up proteomics studies, post-translational modification analysis, and other protein mass spectrometry methods.
You can also check out the two posters we’ll have, both during Wednesday’s poster session.
• Poster #590: “Using Orthogonal Techniques for Protein-Peptide Separation to Generate Comprehensive HDMSe Mass Spectral Libraries from an E. coli Model System”
• Poster #674: “The BluePippin Automated Size-Fractionation System for Proteins”
We hope to see you there!
New Prostate Cancer Research Indicates Rapid Accumulation of DNA Mutations
Researchers studying the behavior of prostate cancer have shed light on how the cancer develops mutations so rapidly. In contrast to other cancers that seem to progress by adding mutations one at a time, the prostate cancer samples that were analyzed showed evidence of a rapid chromosomal scramble resulting in several alterations that disrupt many genes at once. The scientists named this phenomenon “chromoplexy.”
The findings are described in “Punctuated Evolution of Prostate Cancer Genomes,” published in Cell by a large collaboration of scientists with senior authors Francesca Demichelis, Mark Rubin, and Levi Garraway. We were delighted to see that the team used Pippin Prep automated size selection as part of their sample preparation workflow in this impressive research.
For this work, the team sequenced 57 tumor genomes and their matched normal tissue, hoping to build on findings from earlier exome studies and to provide a better understanding of the onset and progression of prostate cancer. They also analyzed copy number changes across the genomes and studied the chromoplexy phenomenon they found, through which large chunks of the genomes had been jumbled in a short period of time. “These complex rearrangement events occur in the majority of prostate cancers and may commonly inactivate multiple tumor-constraining genes in a coordinated fashion,” the authors note.
They add, “Our modeling suggests that chromoplexy may induce considerable genomic derangement over relatively few events in prostate cancer and other neoplasms, supporting a model of punctuated cancer evolution.”
Pippin and PacBio: Better Together
Update: Just announced– Sage Science and Pacific Biosciences have partnered to offer users longer reads than ever. Click here to read the press release.
The new SMRTbellTM 20kb sample prep protocol using the BluePippin is posted on the PacBio® SampleNet site, click here to view the page or download the document. Instructions for using the BluePippin High Pass cassette definition for the SMRTbell protocol can be found here.
Original Post:
Many thanks to all the scientists who stopped by the Sage suite during the Advances in Genome Biology & Technology meeting in Marco Island — we were thrilled to see you! When we weren’t busy handing out glowing medallions,  the Sage Science team had a great time catching up with people, attending the excellent scientific presentations, and walking the halls of the poster sessions.
the Sage Science team had a great time catching up with people, attending the excellent scientific presentations, and walking the halls of the poster sessions.
Along with other attendees, we were really impressed by the posters and presentations showing data from the PacBio® RS. A number of talks referred to the company’s single-molecule, real-time (SMRT®) sequencing, which generates extremely long reads — one scientist said he’d produced a 26 kb read on the PacBio RS. Between the read length and unique base modification detection capability, the sequencing platform is particularly useful for de novo assembly, genome finishing, SNP discovery, and epigenetic analysis.
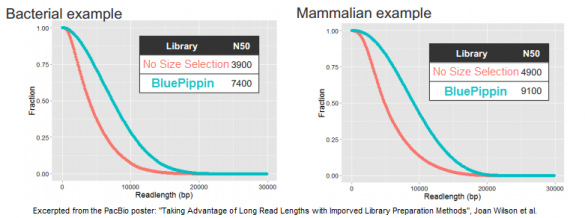
In its quest to produce sequence from even longer DNA fragments, PacBio recently tested our BluePippin platform for size selection; the results of those studies were the basis for a poster we displayed at AGBT. In it, the PacBio authors demonstrate a doubling of the N50 subread length – the value at which 50% of unique DNA bases are in reads longer than this value – when using the BluePippin platform compared to the standard workflow (click to enlarge this excerpt from the poster):
For generating 20 kb libraries that are optimal for the platform’s current polymerase, the authors note that “when combined with size selection to remove the shorter fragments that tend to load more favorably than long fragments, these libraries show greatly increased subread lengths.”
Looking at three examples of libraries for bacterial, fish, and mammalian DNA, the authors report that the N50 subread lengths increased by 90%, 110%, and 86%, respectively. Some of this data was also included in an AGBT plenary presentation by Jonas Korlach, Chief Scientific Officer of PacBio, who cited the BluePippin platform as one of the new improvements for PacBio customers.
Some PacBio customers are already using the BluePippin platform, and we look forward to working with more of them in the coming months. Together, these technologies offer a great and unique solution for truly long-read sequencing.
Pippin Pulse
Field Inversion Gel Electrophoresis
We’ve set up this post to provide Pippin Pulse users with a resource to keep updated on new protocols and to post references that might be useful. We also invite users to feel use this page as a forum, if you see fit, and we’d enjoy showing interesting gel images and data (email these to support@sagescience.com).
Our go-to book on theory:
“Electrophoresis of Large DNA Molecules:Theory and Applications” (E. Lai and B. Birren, eds), Current Communication in Cell & Molecular Biology Vol. 1,. Cold Spring Harbor Laboratory Press, New York, 1990
Download the Pippin Pulse user manual here
Our Gel Set-up
We use a Galileo Model 1214 RapidCastTM Mini Gel Unit running 12 X 14 cm gels using Lonza SeaKem® Gold Agarose. We use our Pippin Tris-TAPS buffers (which you can order directly from us in the US and Canada, Part No. KBB1001, under “Accessories” on our ordering page) or 0.5X TBE. The formulations can be found in the Pippin Pulse user manual or separately here.
Gel images of our 1-50kb and 3-70kb protocols:
(click to enlarge images)