See You at BioConference Live: Genetics and Genomics
This week brings a new experience for us: Sage is participating in a virtual conference! For those of you not yet familiar with it, BioConference Live runs several meetings each year, all 100 percent online. Attendees can tune in for excellent talks, meet company representatives in the exhibit hall, and even pick up giveaways — all from the comfort of home or office.
The Genetics and Genomics conference is being held this week, on August 21-22. We’re really impressed with the speakers at this meeting: keynotes include luminaries George Church, Drew Endy, Mike Snyder, and David Baker. There are also several tracks to choose from in the two-day meeting, such as bioinformatics and quantitative genomics; clinical genomics; epidemiology and population genetics; synthetic and systems biology; and more.
Unlike an in-person conference, you can tune in for just the sessions that interest you, without anyone noticing when you duck out. There’s no travel involved, no need to dress up, and best of all, registration is free. You can sign up here.
Sage Science will have a booth in the exhibit hall, where we’d be happy to meet you and answer any questions about how automated DNA size selection could improve your sequencing workflow. (That sounds strange for a virtual event, but there will be real, live people behind those avatars, ready to chat.) You can find us by searching for “Sage” in the exhibit hall or by looking for our fancy blue booth.
We hope to see you there!
In Sanger Study, BluePippin Contributes to Improved Construction Method for Mate-Pair Libraries
Scientists from the Wellcome Trust Sanger Institute recently published “An improved approach to mate-paired library preparation for Illumina sequencing,” a paper that came out in the open access journal Methods in Next Generation Sequencing.
The publication, from lead author Naomi Park, is the culmination of a project designed to establish an alternative to currently available commercial kits for mate-pair library construction. These existing methods of circularization “have differing limitations and are often linked to a single sequencing platform in a kit format, which may not be cost effective,” the authors write.
Mate-pair sequencing is often used for de novo genome assembly, detection of structural variants, genome finishing, and other projects in which long-range sequence data is beneficial. But preparing these libraries tends to be challenging — and gets even worse when there’s not much DNA to start with or when the DNA is degraded, the scientists note.
The paper compares a new method developed by the Sanger scientists with several existing mate-pair library construction approaches. “In order to generate unbiased and diverse Illumina mate-paired libraries containing even genomic sequence either side of a common adapter sequence, we altered the Illumina mate-pair protocol to use a modified SOLiD 4 hybridisation and ligation circularisation approach,” they write. Tests involving multiple libraries and sample types demonstrated that the new method increases library complexity and quality while reducing chimeras.
Most of the libraries involved in this study were size selected with BluePippin. “Alternative methods of size selection are amenable to this method, but may result in a differing breadth of size range and/or recovery,” the authors write. After comparing results from Pippin, manual gels, and bead size selection, they conclude, “The Blue Pippin provides the greatest recovery and accuracy of currently available commercial methods.”
BluePippin Bulletin: Size Selection of 40kb and 50kb DNA
Well, we’ve finally gone where no automated DNA size selection provider has gone before. Using compression-busting pulse-field programs and fancy gel formulations, we’ve tackled the large cut.
Our latest cassette type, BHF7510, comes with four cassette definitions:
1. High Range 40kb Marker W1- Tight
2. High Range 40kb Marker W1- Broad
3. High Range 50kb Marker W1- Tight
4. High Range 50kb Marker W1- Broad
In all four cases, the size selection is centered on the 40kb or 50kb target; range mode selections are not allowed. The “Tight” definitions provide very narrow distributions, while the “Broad” definition doubles the duration of elution to provide higher yields. Below is a summary of some our validation data.
Thanks to our crack R&D team for unlocking the mysteries of HMW fragments!
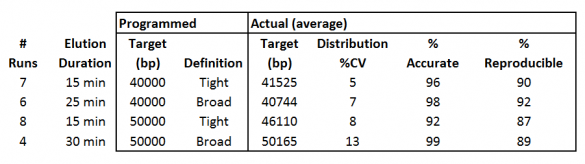
Figure 1. Data summary from High-Range cassette validation tests. Eluted DNA was run on agarose gels using the Pippin Pulse system (3-70kb preset program) and analyzed with the TL100 analysis software from Totallab, Ltd.
From the Trenches: PacBio User Shares Experience with BluePippin
As the BluePippin makes its way to more users of the Pacific Biosciences sequencing platform, there is great data emerging on how our high-pass size selection protocol really complements the sequencer.
A particularly nice example of this comes from Lex Nederbragt, research fellow at the University of Oslo and member of the Norwegian High-Throughput Sequencing Centre, who recently put up a blog post about his experiences with BluePippin and PacBio. He includes plenty of data showing sequencing runs with and without BluePippin selection.
Nederbragt writes that BluePippin was very effective at reducing small fragments from the library, boosting average read lengths and longest subread lengths, and shifting base distribution significantly to the longest reads. Be sure to check out the figures in his blog post for specific data.
Many thanks to the Norwegian High-Throughput Sequencing Centre for their work with BluePippin and to Nederbragt for sharing this information!
Back from Baltimore and the Stellar PacBio UGM
Earlier this week Pacific Biosciences hosted a user group meeting at the University of Maryland in Baltimore, and we were pleased to participate in and co-sponsor the event. As our blog readers know, we recently announced a co-marketing partnership with PacBio to provide BluePippin size selection to PacBio users to help them generate longer average read lengths.
At the user group meeting, about 100 attendees came to discuss their own experiences with the sequencing platform and to learn how other people were pushing the boundaries on read length, DNA input requirements, and more. It was great to see so much enthusiasm for PacBio’s sequencer. This is a really engaged community — almost every presentation was followed by several questions and often lively discussion within the audience as people shared their own methods or results.
A key theme at the meeting revolved around the dramatic improvements in throughput and read length since the PacBio system first launched. Speakers got in the habit of noting what year certain data had been generated as shorthand to help attendees understand how the data might look today if the experiment were run with newer enzymes, reagents, or hardware. A speaker from Baylor noted that since receiving their instrument, throughput has increased more than 10-fold while average read length has jumped from 1.5 Kb to 7 Kb. Here at Sage, we are proud to be part of that growth in read length — and we were glad to see speakers reporting a significant boost in average read length with the incorporation of BluePippin into the workflow.
With these extraordinarily long reads, many of the speakers presented information on genetic elements or attributes that have never been seen before, including connections of distant tandem repeats, genome-wide methylation in pathogens, and fully sequenced mRNA transcripts. There was also quite a bit of excitement around moving toward de novo mammalian sequencing with the PacBio workflow, which can currently perform de novo sequencing and automated assembly/finishing of microbial genomes.
From Sage Science, Chris Boles offered an update on BluePippin for PacBio. (For the current protocol, click here.) He presented data from one genome center showing a project in which the N50 median read length of 4,800 bases was boosted to 9,100 bases by adding BluePippin to remove smaller fragments from the library. Boles also reported that we are working to develop additional protocols with PacBio to keep extending those N50 numbers. We look forward to continuing to help PacBio users generate even more impressive read lengths from their sequencers!






