In Skin Microbiome Study, Scientists Find Host Genotype Influences Bacterial Populations
A paper that came out in the Journal of Innate Immunity this summer reports that a person’s genotype affects which microbes will colonize his or her skin, which in turn may alter that person’s defense mechanisms against pathogenic organisms.
“Skin Microbiome Imbalance in Patients with STAT1/STAT3 Defects Impairs Innate Host Defense Responses” comes from a team of scientists in Boston and The Netherlands. Lead author Sanne Smeekens, from Radboud University Nijmegen Medical Center, and her collaborators investigated patients with immunodeficiencies linked to mutations in STAT1 and STAT3 that increase susceptibility to skin and mucosal infections, particularly from fungal pathogens or Staphylococcus aureus.
To determine the implications of these mutations in microbiome and host defense, the scientists compared skin and oral samples from several patients with age-matched healthy controls. Microbial colonies were assessed with 16S rRNA sequencing, performed on Illumina MiSeq after size selection with Pippin Prep.
The team found that immunodeficient patients’ microbiomes contained more Gram-negative bacteria (particularly Acinetobacter) and less Corynebacterium than their healthy counterparts. Functional studies revealed that the difference in microbiome composition leads to an inhibited immune response to Candida albicans and S. aureus in these patients.
“These data in patients with immunodeficiencies prove that the microbiome can influence host defense and could open the possibility of microbiome-based adjuvant therapy in patients with immune defects,” the authors conclude.
All Conferenced Out: Great Experiences at NGx and BioConference Live
We’ve just come off a busy week of conferences! In addition to BioConference Live: Genetics and Genomics, our first virtual scientific meeting, we also attended NGx: Applying Next-Generation Sequencing in Providence, Rhode Island.
NGx, one of the best-known next-gen sequencing events, had stellar keynote speakers, including Yaniv Erlich from Whitehead Biomedical Research Institute and Jeffery Schloss from NHGRI. We were eager to speak with attendees at our booth and to hear the talks; with this meeting, we always get a great glimpse of where the NGS field is. Scientists from major genome centers and smaller labs alike spoke about how they’re keeping up with rapid technology changes and implementing best practices.
One comment that really struck us came from Stuart Brown at New York University School of Medicine. In a talk entitled “Can We Maintain Sanity as NGS Pipelines Change?” Brown noted that in planning a two-year sequencing project, scientists can no longer expect to use the same sequencing kits and bioinformatics solution for the entire project. It’s a great point and one that we think about a lot as we strive to make sure that our size selection tools work seamlessly with each version of all commercially available sequencing platforms.
As the NGx show wrapped up, our experiment with a 100% online conference began. BioConference Live: Genetics and Genomics kicked off bright and early, but the talks are saved on the website and made available on-demand through free registration. Keynoters George Church and Mike Snyder got rave reviews on Twitter (though, as we discovered, that doesn’t really compare to getting a pastry and exchanging reviews with other attendees during a coffee break).
Compelling talk titles were essential in this type of venue. Drew Endy’s keynote presentation on “Aliens, Computers, and Engineering Biology” got attention; others that we found intriguing were “You Can and Must Understand Synthetic Biology” (Andrew Hessel) and “Genome Hacking” (the very popular Yaniv Erlich). We were thrilled to find people visiting our booth — thanks to all the other virtual experimenters who stopped by.
While we really enjoyed this virtual conference, we’re not giving up the old-school, in-person meetings just yet. We missed catching up with our colleagues, the obligatory buffet lunch, and the chance to sneak out of a session for a quick walk on the beach (we’re looking at you, Marco Island). But mingling via our avatar was certainly a new experience!
See You at BioConference Live: Genetics and Genomics
This week brings a new experience for us: Sage is participating in a virtual conference! For those of you not yet familiar with it, BioConference Live runs several meetings each year, all 100 percent online. Attendees can tune in for excellent talks, meet company representatives in the exhibit hall, and even pick up giveaways — all from the comfort of home or office.
The Genetics and Genomics conference is being held this week, on August 21-22. We’re really impressed with the speakers at this meeting: keynotes include luminaries George Church, Drew Endy, Mike Snyder, and David Baker. There are also several tracks to choose from in the two-day meeting, such as bioinformatics and quantitative genomics; clinical genomics; epidemiology and population genetics; synthetic and systems biology; and more.
Unlike an in-person conference, you can tune in for just the sessions that interest you, without anyone noticing when you duck out. There’s no travel involved, no need to dress up, and best of all, registration is free. You can sign up here.
Sage Science will have a booth in the exhibit hall, where we’d be happy to meet you and answer any questions about how automated DNA size selection could improve your sequencing workflow. (That sounds strange for a virtual event, but there will be real, live people behind those avatars, ready to chat.) You can find us by searching for “Sage” in the exhibit hall or by looking for our fancy blue booth.
We hope to see you there!
In Sanger Study, BluePippin Contributes to Improved Construction Method for Mate-Pair Libraries
Scientists from the Wellcome Trust Sanger Institute recently published “An improved approach to mate-paired library preparation for Illumina sequencing,” a paper that came out in the open access journal Methods in Next Generation Sequencing.
The publication, from lead author Naomi Park, is the culmination of a project designed to establish an alternative to currently available commercial kits for mate-pair library construction. These existing methods of circularization “have differing limitations and are often linked to a single sequencing platform in a kit format, which may not be cost effective,” the authors write.
Mate-pair sequencing is often used for de novo genome assembly, detection of structural variants, genome finishing, and other projects in which long-range sequence data is beneficial. But preparing these libraries tends to be challenging — and gets even worse when there’s not much DNA to start with or when the DNA is degraded, the scientists note.
The paper compares a new method developed by the Sanger scientists with several existing mate-pair library construction approaches. “In order to generate unbiased and diverse Illumina mate-paired libraries containing even genomic sequence either side of a common adapter sequence, we altered the Illumina mate-pair protocol to use a modified SOLiD 4 hybridisation and ligation circularisation approach,” they write. Tests involving multiple libraries and sample types demonstrated that the new method increases library complexity and quality while reducing chimeras.
Most of the libraries involved in this study were size selected with BluePippin. “Alternative methods of size selection are amenable to this method, but may result in a differing breadth of size range and/or recovery,” the authors write. After comparing results from Pippin, manual gels, and bead size selection, they conclude, “The Blue Pippin provides the greatest recovery and accuracy of currently available commercial methods.”
BluePippin Bulletin: Size Selection of 40kb and 50kb DNA
Well, we’ve finally gone where no automated DNA size selection provider has gone before. Using compression-busting pulse-field programs and fancy gel formulations, we’ve tackled the large cut.
Our latest cassette type, BHF7510, comes with four cassette definitions:
1. High Range 40kb Marker W1- Tight
2. High Range 40kb Marker W1- Broad
3. High Range 50kb Marker W1- Tight
4. High Range 50kb Marker W1- Broad
In all four cases, the size selection is centered on the 40kb or 50kb target; range mode selections are not allowed. The “Tight” definitions provide very narrow distributions, while the “Broad” definition doubles the duration of elution to provide higher yields. Below is a summary of some our validation data.
Thanks to our crack R&D team for unlocking the mysteries of HMW fragments!
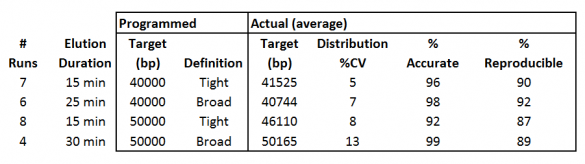
Figure 1. Data summary from High-Range cassette validation tests. Eluted DNA was run on agarose gels using the Pippin Pulse system (3-70kb preset program) and analyzed with the TL100 analysis software from Totallab, Ltd.





