It’s Baaack! Festival of Genomics Returns to Beantown
It’s time for a genomics party! The Festival of Genomics returns to Boston next week, and we’re already looking forward to it. Sage is proud to be a sponsor and exhibitor at this great event, which brings together more than 1,000 people to share the latest in great science and clinical impact.
The agenda is jam-packed with great speakers, and we’re delighted to see that this year’s event includes a whole track for rare disease patients. General sessions will cover large-scale initiatives — such as organizing 100,000 human genome assemblies, building a human cell atlas, and improving diversity in population studies — as well as relevant drug discovery efforts, CRISPR/Cas9 advances, oncology studies, and much more. We are particularly eager to hear from Zivana Tezak, who will speak about the FDA’s precision medicine strategy.
As always, there will be lots of opportunities to hear about how various technologies are being deployed to improve scientific results. To learn more about automated DNA size selection and how it can make a difference in your lab, check out our booth near Horizon Stage 2. We hope to see you there!
What’s the difference between the 4-10 Kb vs2 and 6-10 Kb vs3 high pass protocols?
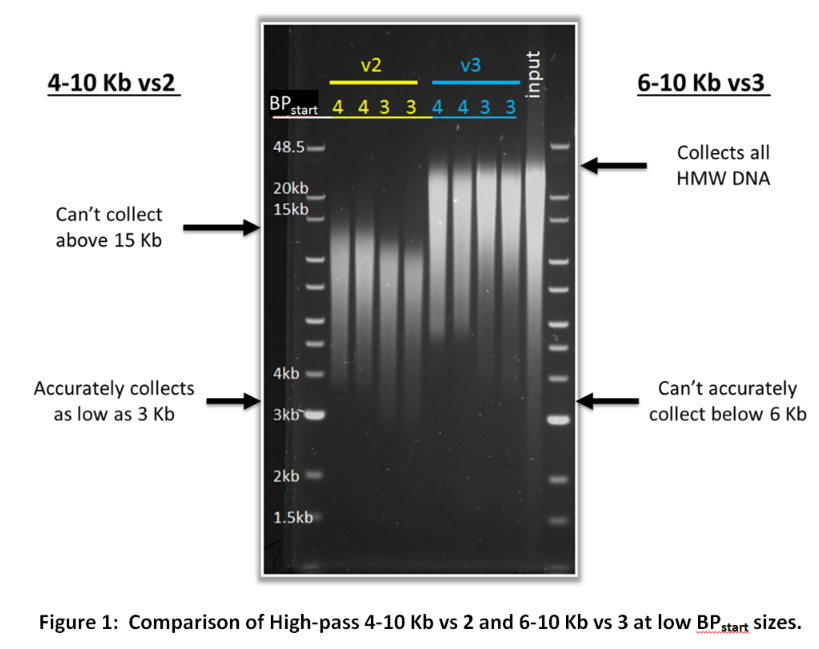
Customers may have noticed two very similar cassette definitions; 0.75%DF Marker S1 high-pass 4-10kb vs2 and 0.75%DF Marker S1 High-Pass 6-10kb vs3. Since these two protocols cover a similar range, they might seem a bit redundant. However, both of these cassette definitions are important because they can complement the limitations of the other. The 6-10 Kb vs3 protocol cannot start collection below 6 Kb accurately while the 4-10 Kb vs2 protocol can accurately begin collection in the 4-6 Kb range (and can even achieve reasonable accuracy as low as 3 Kb). However, the 4-10 Kb vs2 protocol can only collect up to ~15 Kb DNA fragments while the 6-10 Kb vs3 protocol will collect all high molecular weight DNA. The figure below demonstrates the limitations and strengths of these two protocols.
If you are unsure whether to use the 4-10 Kb vs 2 or the 6-10 Kb vs 3 cassette definition for your high pass filtering, consider the criteria below:
- If you are following the protocol produced by one of our partners (such as Pacific Biosciences) use the cassette definitions they recommend.
- If you want to start collection between 6-10 Kb, use the 6-10 Kb vs 3 cassette definition.
- If you want to start collection between 4-6 Kb and you don’t need DNA over 15 Kb, use the 4-10 Kb cassette definition.
- If you want to start collection between 4-6 Kb and you absolutely need DNA over 15 KB, contact customer service for assistance.
Hopefully this post clears up some of the confusion customers have had with these two protocols. For more information on programming a high pass protocol on the Blue Pippin, please refer to this user guide.
Scientists Compare PacBio, Oxford Nanopore Transcriptome Results
We enjoy a good technology evaluation as much as the next scientist, particularly when it comes to sequencing. So we were quite interested in a recent F1000Research publication about long-read sequencing platforms from researchers at the University of Iowa, the University of Oxford, and other institutions.
From senior author Kin Fai Au and collaborators, “Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis” presents a nice assessment of the pros and cons of long-read sequencing tools. The authors note that PacBio libraries were prepared using SageELF size selection, while the Oxford Nanopore libraries were not size-selected. Some of the study results can be explained by the difference in sample prep.
To compare the technologies, scientists sequenced the transcriptomes of human embryonic stem cells with PacBio, ONT, and Illumina (short reads were used for comparison purposes as well as for building hybrid assemblies). They note that long reads have been especially informative for transcriptome studies, including gene isoform identification.
In this analysis, both platforms were able “to provide precise and complete isoform identification” for a small library of known spike-in variants and for more complex transcriptomes as well. “PacBio has a slightly better overall performance, such as discovery of transcriptome complexity and sensitive identification of isoforms,” the team reports.
Delving into details, the long-read platforms performed similarly in read length, based on a comparison of mappable length. The team found that a higher proportion of PacBio reads could be aligned to reference genomes compared to ONT reads. Throughput was also noticeably different: “the yield per flow cell of ONT is much higher than PacBio, because each nanopore can sequence multiple molecules, while the wells of PacBio SMRT cells are not reusable,” the authors note. Error rate was another area of divergent results. PacBio CCS reads had an error rate “as low as 1.72%,” the scientists report, giving data from that platform “higher base quality than corresponding ONT data.”
“PacBio can generate extremely-low-error-rate data for high-resolution studies, which is not feasible for ONT,” the scientists add, noting that ONT advantages include high throughput and lower expense. “The cost for our ONT data generation was 1,000–2,000USD,” they report.
In addition, the scientists assessed hybrid approaches for both platforms, adding short Illumina reads for error correction. It was the first known pairing of ONT and Illumina reads for this purpose. “As this first use of ONT reads in a Hybrid-Seq analysis has shown, both PacBio and ONT can benefit from a combined Illumina strategy,” the team writes. The authors note that both long-read sequencing tools have improved significantly with recent models and predict that future enhancements will be a boon to transcriptome studies as well.
Podcast: Nanopore Expert Mark Akeson on Challenges and Opportunities in Sequencing
Mark Akeson knows a thing or two about perseverance. After spending years as a soil biologist in Guatemala where he endured a series of parasite infections, he went on to become a pioneer in nanopore sequencing technology, which he has been developing for more than 20 years even though highly regarded scientists insisted it would never work.
His story is the subject of an interview with Mendelspod host Theral Timpson, and the conversation — which took place earlier this year — provides fascinating insight into the world of nanopore sequencing.
Now co-director of the biophysics laboratory at the University of California, Santa Cruz, and a consultant to Oxford Nanopore, Akeson told Timpson that current nanopore technology has a number of challenges but also has plenty of room for significant improvement. Right now, accuracy of single-pass reads is still lower than that of short-read sequencing platforms, and sample prep issues limit the length of fragments that can be fed through the pore. But having already overcome major obstacles — finding the right size sensor, improving sensitivity, and moving DNA reliably one nucleotide at a time — Akeson is confident the technology will get even better.
As he sees it, the major advantage of nanopore sequencing is the ability to interpret DNA or RNA directly from the cell. “You’re reading what nature put there, not only the bases but modifications,” he said. In addition, nanopores allow for very long reads (Akeson said his lab is routinely generating 200 Kb sequences) and are ideally suited for sequencing in the field.
This work may not have been possible without the commitment of program officers at NHGRI, whom Akeson praised as “visionaries on this research.” Now that the approach has finally been proven to work, “the whole nanopore sequencing endeavor is going exponentially faster,” he added.
Looking ahead, Akeson predicted that 20 years from now genome sequencing will be so cheap it’ll just be given away to consumers, with profits coming from follow-on interpretation services. One day, he said, we’ll look back on the $1,000 genome or even the $100 genome as very expensive. Sequencing technology will continue to improve over time because it’s “too important a technology to say ‘We’re done,’” he added, noting that accuracy, read length, and throughput will be areas of focus in the coming years.
Nabsys Returns: CEO Barrett Bready on the Importance of Structural Variation
A new podcast from Mendelspod features an interesting interview with Barrett Bready, CEO of electronic mapping firm Nabsys, who emphasizes the growing need to incorporate structural variation data into genome studies.
In the discussion, Bready describes his company’s platform, which relies on voltage-powered, solid-state nanodetectors to generate map-level information. Each nanodetector can cover 1 million bases per second, Bready said, and can be multiplexed for a highly scalable system. It’s a “really high-speed, highly scalable way of getting structural information,” he added.
Bready noted that the genomics community has realized the need for long-range information, estimating that known structural variants now make up about 60 Mb of the human genome, a number that has increased rapidly in the last few years even as the amount of sequence attributed to single-nucleotide variants has stayed the same. Nabsys aims to democratize access to structural information by producing a cost-effective mapping tool for routine analysis of these large variants.
This information will complement short-read data, Bready said, which necessarily sacrifices assembly contiguity due to the need to cut DNA into small fragments prior to sequencing. The Nabsys platform works with high molecular weight DNA to capture extremely long-range information. He also said that electronic mapping data offers more value than optical mapping technologies.
Beta testing for the new platform is expected to begin early next year.