BluePippin “Mid-Range” Size Selection: 18-27 kb
We’re pleased to be shipping our Mid-Range size selection cassette (BMF7510) for the BluePippin. While our Low-Range cassette (BLF7510, 1kb-10kb) works for many mate-pair library construction protocols, we’ve been hearing from our customers that there is interest in creating larger circularized molecules. This cassette, which continues to extend the range and flexibility of the BluePippin product line, should serve as a useful tool for large-fragment preps and help produce more diverse and meaningful data.
(Some internally generated validation data can be found on this pdf.)
In this blog, we’d like to outline some characteristics on the product to help users develop appropriate methods.
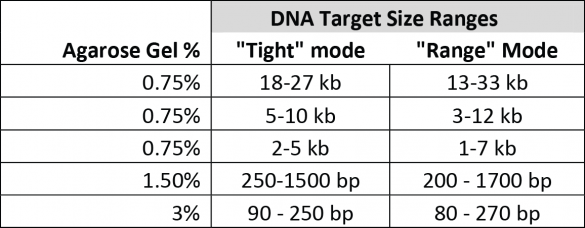
CVs and target ranges
We define all target ranges in terms of what a user can accurately collect by entering a value in “tight” mode using our software. We also have a specification, “Minimum Size Distribution as Expressed by CV,” that describes the minimum distribution of fragments that may be collected given the capabilities of the system and the resolution of the gel. For instance, as a rule of thumb, a CV of 8% (our spec for smaller targets) will yield a range of fragments that are at most + 16% of the median fragment. However, the mid-range targets collect at a comparatively larger distribution minimum (20% CV), which limits the effective range of accurate “tight” cuts. This chart compares the accurately selectable “tight mode” ranges to the “range mode” ranges for the gel cassette calibrations available for the BluePippin at this time:
Sample Load Dependence
To calibrate the mid-range cassettes, we use a 5 ug sample load of sheared E. coli genomic DNA. Higher sample loads (up to 10ug) will usually run somewhat slower, relative to the marker DNA, and for this reason, you will need to program higher bp target or range values in the protocol to collect a desired size fraction. In our tests, when using a 10 ug input load, bp target values should be increased by 10-15% to compensate for the changes in mobility caused by the increased load.
Another factor to consider is the size distribution of the input DNA. For calibration and testing, we use an E. coli genomic DNA sample that has a very broad, almost flat size distribution. Using such samples simplifies our calibration process, which involves accurately sizing fractions sliced from the flat input samples. However, most input samples for mate-pair libraries are generated by methods that produce much narrower size distribution (DigiLab Hydroshear, for instance). Such samples will therefore show a different mobility dependence on input load than our E. coli genomic DNA test samples. For this reason, customers should plan on conducting some pilot experiments on non-valuable samples to investigate the mobility-load relationship produced by their specific library protocol.
Bottom line on 0.75% Mid-range cassette definition
- BluePippin 0.75% mid-range cassettes facilitate size fractionations of DNA samples out to 30kb.
- The CVs of tight size selections will be wider than those obtained from other BluePippin and Pippin Prep cassettes (mid-range CVs around 20%).
- The mid-range protocols show a significant mobility dependence on input load. Sage uses a 5 ug genomic sample with a broad size distribution for calibration. Higher input loads and samples with narrow size distributions will require user optimization of programmed size values for accurate results.
- Input loads higher than 10 ug per lane will have unpredictable results, and are not recommended.
If you plan to use the the BluePippin for mate-pair library construction, we’d love to hear of your progress or suggestions. To share, please contact us at info@sagescience.com or reply to this blog.
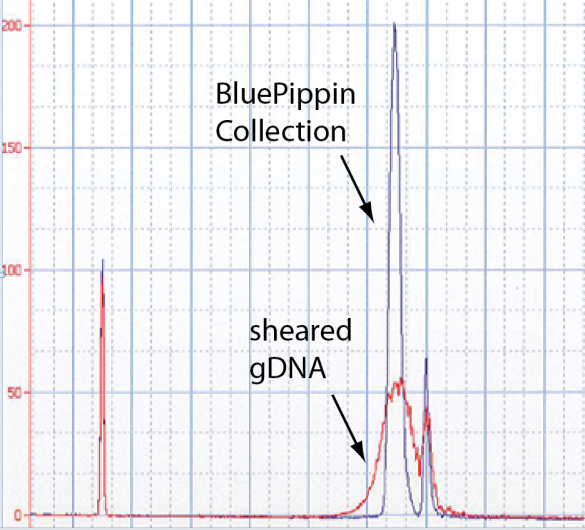
The image below is an Agilent Bioanalyzer trace of a nice 8 kb collection in the middle of a gDNA shear. This is near the limit of the Bioanalyzer’s detection target range.
Publication: Spike-In Standards for Managing Systematic Sequencing Errors
Here’s a paper worth checking out: “Synthetic Spike-in Standards Improve Run-Specific Systematic Error Analysis for DNA and RNA Sequencing” from lead author Justin Zook at the National Institute of Standards and Technology. Published in PLoS One last month, the paper describes a study of ways to better manage systematic errors in DNA or RNA sequencing.
Most sequencing work currently relies on algorithms that recalibrate base scores after calculating a correction factor using either a subset of the sequenced data set or a separate data set, the paper’s authors write. They propose using synthetic spike-in standards, in this demonstration using RNA spike-ins for sequencing human RNA. This is followed up with base recalibration with the Genome Analysis Toolkit (GATK from the Broad Institute) that more accurately adjusts based on the spike-in’s unique sequence signature.
“Compared to conventional GATK recalibration that uses reads mapped to the genome, spike-ins improve the accuracy of Illumina base quality scores by a mean of 5 Phred-scaled quality score units, and by as much as 13 units at CpG sites,” the authors write. “In addition, since the spike-in data used for recalibration are independent of the genome being sequenced, our method allows run-specific recalibration even for the many species without a comprehensive and accurate SNP database.”
In a paper that focuses on improving quality and uniformity, we were delighted to see that our Pippin platform was used for the cDNA size selection step with Illumina sequencing.
Congratulations to authors Justin Zook, Daniel Samarov, Jennifer McDaniel, Shurjo Sen, and Marc Salit!
For NRC Team, Pippin Platform Leads to Higher-Quality Assemblies
At the DNA Technologies Laboratory at the National Research Council of Canada, scientists are using the Pippin size selection platform to improve the quality of their genome assemblies.
Andrew Sharpe, Research Officer and Group Leader of the Saskatoon-based laboratory, got his first Pippin Prep last year. He added a second Pippin Prep as well as the longer-fragment Blue Pippin to his arsenal earlier this year.
In Sharpe’s lab, which also serves as a core facility for NRC and other Canadian government agencies, assembly projects tend to focus on large plant and fungal genomes. His team relies on Illumina and 454 sequencing, often adopting a hybrid assembly approach to take advantage of both platforms.
“The majority of libraries going through are the shorter, standard pair-end libraries of 200 to 400 bases,” Sharpe says, noting that those libraries run on all three Pippin machines. Longer mate libraries — usually in the range of 3kb to 10 kb — are also a good fit for the Pippin, he adds.
Sharpe and his colleagues use the Pippin platform to create multiple pair-end libraries for the same sample — constructing, for instance, three libraries with 200-base, 300-base, and 400-base inserts — and then assemble all of those sequences together, often using SOAPdenovo. “If you assemble one of the libraries, then you’ll end up with an assembly. But if you assemble all three together using three different lengths, you get quite a bit better product,” Sharpe says. “The nice thing with the Pippin Prep is being able to easily get those discrete size ranges.”
Before Sharpe had the Pippin, his team spent a lot of time on manual gel extractions. “Having the Pippin makes things quite a lot more efficient on the labor side,” he says. Now he’s looking to the Blue Pippin to take over for the field inversion gel electrophoresis his team runs for making larger 454 mate libraries. “The Blue Pippin offers the prospect of actually speeding up that process and hopefully getting away with less amounts of DNA,” Sharpe says. “You normally need a lot of DNA to operate on the standard FIGE gel, but with the Blue Pippin we should be able to get away with less.”
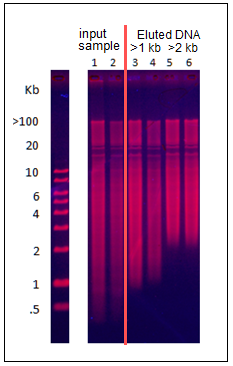
BluePippin:Collecting all DNA “Greater Than” 1kb
We have received several requests to use the Pippin to collect all remaining DNA above a programmed base pair value from a sample. Although Pippins currently have the capability to collect all DNA after a run time threshold (using the “Time” programming mode), there is no method to elute the entire sample after a programmed base pair value . Also, our protocol editor requires users to enter an ending base pair value (BP End) in the “Range” mode and will not accept values above 50,000 bp.
For the BluePippin, we have developed a protocol for this purpose, and named the cassette file “0.75% Greater Than – Marker S1”. This requires our 0.75% dye-free gel cassettes kit for lower ranges (BLF7510). With this protocol, users enter a 4 hour run time, and enter a 50,000 bp value into the “BP End” field using the “Range” programming mode. The 50,000 is a dummy value that tells the instrument to continue collecting until the end of the run.
At this time, the “0.75% Greater Than – Marker S1” cassette file is not available in the standard menu of cassette types, but we can provide it to you separately. Contact us if you are interested.
Broad Institute Teams with Sage Science for Automated Sizing
Don’t miss this great blog post from the Broad Institute (“A Sage partnership”) describing collaborative work between their genome sequencing team and Sage Science to design a better size selection process for the Broad’s sequencing pipeline.
Headed up by Sheila Fisher, assistant director of technology development for the Broad’s Genome Sequencing Platform, the goal was to replace error-prone, tedious manual gel extractions in the sample prep workflow. Working with Sage’s Pippin platform, Sheila and her team were able to automate the size selection step, improving accuracy and eliminating the chance for cross-sample contamination.
An added bonus was that Pippin sizing offered much higher yields than manual gel extraction had, allowing Fisher’s team to accept samples with just 100 nanograms of DNA, instead of the 3 or 4 micrograms the pipeline originally required. “This opened up a significant number of samples to the process that we couldn’t sequence before,” says Sheila in the blog post. “We were able to build a very strong partnership with Sage, and the result was a true co-development project.”
We couldn’t have said it better ourselves. It’s truly a pleasure to continue with our great collaboration with the Broadies!