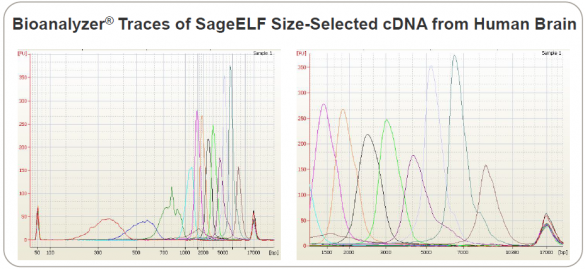
Poster on Full-Length Transcript Sequencing Includes SageELF
We are pleased to report the first customer poster featuring the newest addition to our product lineup, the SageELF whole-sample fractionation tool. “Single Molecule, Real-Time Sequencing of Full-length cDNA Transcripts Uncovers Novel Alternatively Spliced Isoforms” comes from scientists at Pacific Biosciences and the University of Washington.
The poster describes a human gene expression study in which the scientists were able to generate full-length isoform sequences, with some transcripts longer than 10 Kb. Isoform sequencing was conducted using the PacBio® RS II DNA Sequencing System. The cDNA sequences came from the MCF-7 human breast cancer cell line as well as from brain, heart, and liver cells.
The authors note: “Even in extensively profiled sample types, the method has been able to uncover large numbers of novel alternatively spliced isoforms and previously unannotated genes.” This ability to produce full-length transcript sequences offers a unique way to examine alternative splicing in eukaryotic organisms.
The team used SageELF, an automated sample prep device that generates 12 contiguous fractions from a single DNA or protein sample. It can be used in NGS workflows to build libraries with multiple insert sizes from the same sample, as well as to preserve precious samples. This Bioanalyzer trace of SageELF fractions is excerpted from the scientific poster:
Illumina Workflow: Pippin or SageELF for Mate-Pair Sequencing
Most Sage customers use their Pippins with an Illumina workflow, so for the next few weeks we’ll be taking a look at some of the most common or interesting applications of automated DNA sizing with the HiSeq, MiSeq, and even the GA II or GA IIx workhorses.
Today we look at mate-pair sequencing, a protocol using a large insert between reads to cover greater genomic distances. The approach is used to span highly repetitive regions and can result in longer contigs and fewer assembly gaps. Often, mate-pair sequence data is combined with shorter-insert paired-end reads to cover a genome more thoroughly. It is used for de novo sequencing as well as structural variant detection and genome finishing.
For Illumina sequencing, the Pippin Prep and BluePippin save time and provide a reproducible alternative to manual size selection that will ensure better sequencing results. Mate-pair sequencing can be tricky, so the more precise your library sizing, the more accurate your data will be in the end. If you’re performing the newer mate-pair sequencing with Nextera, Illumina recommends using the Pippin platform to get “more stringent” sizing than can be accomplished with AMPure alone. (We’re under Size Selection in Chapter 3, beginning on page 40 of the guide.) The document reports that “in our experience running a standard agarose gel does not provide as robust and reproducible results as the Sage Pippin Prep.” We’re honored to be in the official guidelines!
A paper from scientists at the Wellcome Trust Sanger Institute last year offers another take on the mate-pair protocol. The publication, “An improved approach to mate-paired library preparation for Illumina sequencing,” describes optimized techniques to boost library complexity and quality while reducing the occurrence of chimeras. The technique was designed to improve mate-pair success when not much DNA is available or the sample is degraded. BluePippin is a critical part of the final workflow. While the authors noted that other size selection methods could be used, they note, “The Blue Pippin provides the greatest recovery and accuracy of currently available commercial methods.”
Mate-pair sequencing is a great fit for our newest instrument, the SageELF, which performs whole-sample fractionation. SageELF generates 12 contiguous fractions from a DNA sample, allowing scientists to build short-insert and long-insert libraries from the same sample.
Our blog series will continue with peeks into how scientists are using Pippin and Illumina sequencers together for microRNA studies, ChIP-seq, ddRADseq, and more. Check back soon for more!
ASMS in Review: Proteomics and Posters Galore
The Sage Science team headed back to our home base of Beverly, Mass., after four terrific days at the ASMS conference in Baltimore. The scientific content of the show was so good that we didn’t even mind enduring the heat wave that engulfed the city.
We spent a great deal of time absorbing the hundreds of posters presented during the meeting. There were some really impressive scientific efforts, ranging from the very large — such as cataloging the human proteome — to the very small, including nanomaterials.
A poster that caught our interest came from scientists at the Technical University of Munich and other organizations. It described Proteomics DB, a public database containing more than 90 percent of human proteins. (For more in-depth info, check out this article from The Scientist.) This draft of the human proteome, including novel proteins from supposedly noncoding portions of the genome, was generated by compiling existing mass spec libraries along with newly created libraries based on dozens of tissue types, sera, and cell lines. We are real fans of the concept of creating these comprehensive databases, and are proud that building a similar resource for E. coli was the first external use of our SageELF tool.
Another poster highlighted work from the University of Texas and MD Anderson, which demonstrated the use of mass spec together with RNA-seq to get a multidimensional view of cancer stem cells. And in separate work similar to the SDS removal tool we saw earlier in the week, scientists from Dalhousie University in Nova Scotia are developing an interesting technology using plastic columns to precipitate proteins on a Teflon disc.
Thanks again to all of the attendees who stopped by our booth, and to the scientists who helped us learn more about mass spec in a week than we thought possible. See you next year at ASMS!
At PacBio’s UGM, Top-Tier Science and Praise for BluePippin
It was a pleasure to co-sponsor the Pacific Biosciences user group meeting in Baltimore this week. Based on our participation at the same event last year, we had high expectations for it — but even these were surpassed by the quality of speakers, number of attendees, and excellent presentations.
Naturally, the vast majority of talks focused on using the uniquely long reads generated by Single Molecule, Real-Time (SMRT®) Sequencing to do all sorts of interesting projects that can’t be accomplished with short reads. Luke Tallon from the University of Maryland and Adam Phillippy from the National Biodefense Analysis and Countermeasures Center spoke about microbial genomics, Cold Spring Harbor’s Dick McCombie presented work in yeast and plants, and human genome studies were reported by Ali Bashir at the Icahn Institute for Genomics and Multiscale Biology and Tina Graves-Lindsay at Washington University. Other speakers covered more recent applications of SMRT Sequencing, such as metagenomics, full-length isoform sequencing, methylation analysis, and more.
At this meeting last year, we were delighted to have a couple of speakers mention our automated BluePippin size selection tool, which can be used with the PacBio platform to filter out smaller DNA fragments; this allows the sequencer to focus on long fragments, increasing efficiency and average read length. This year, it was a tremendous honor to find that nearly every speaker mentioned using BluePippin. We had that and the new SageELF whole-sample fractionation tool on display at the user group meeting, and we were bowled over by how many attendees came by to check them out. This group kept us on our toes!
It was also great to hear that BluePippin users are already anticipating how they might use the SageELF in their SMRT Sequencing workflows. Some speakers mentioned the possibility that whole-sample fractionation could be used to capture plasmids and other small parts of an organism’s accessory genome that could otherwise be filtered out by the high-pass protocol. We’re eager to see how that and other applications could make these remarkable research efforts even better.
ASMS Kicks Off, Honoring Mass Spec Luminary and Showcasing Cutting-Edge Proteomics
ASMS is underway here in Baltimore, Md., and the meeting has been so engaging that we haven’t even sneaked out to try some local crab cakes. The proteomics conference officially kicked off last night with a talk from Jason Kalirai from the Telescope Science Institute. The astrobiology- and astrophysics-rich presentation was out of the norm for this crowd, and had a palpable cool factor among mass spec scientists.
The first full day was Monday, featuring a program chock-full of concurrent podium sessions, workshops, and a poster viewing. As usual, life science is the best-represented area of mass spec use at this event, with session topics ranging from protein informatics and antibody development to biologic characterization and biomedical applications. Today’s award lecture came from proteomics pioneer Richard Caprioli, who is being recognized for developing MALDI imaging mass spectrometry.
Our team has really enjoyed the unusual setup in the exhibit hall, where we’re stationed in booth #67. As always, the ASMS exhibit hall is large enough to make attendees wish they had golf carts to get around, but this year the exhibitors and posters are interspersed in little islands, making it really easy to sample some of everything (including hot pretzels!). It’s nice being so close to posters that we can dash over and soak in some of the great science at this show.
There are hundreds of posters on display each day, so even with our wandering we only got to see a small fraction of what was available on Monday. One poster we found particularly interesting came from Neil Kelleher’s lab at Northwestern University. It focused on finding new ways to remove SDS, which is an important factor to our new SageELF workflow so it’s something our R&D team has been working on as well.
Speaking of which, if you haven’t stopped by to see the SageELF, please do so. We’ve had terrific conversations with the many attendees who have come to find out how this automated electrophoresis tool can reduce sample complexity, and we look forward to even more as the conference continues.
And now, if you’ll excuse us, all eyes are on the legendary hospitality suites tonight. After a long day at the conference, we’re looking forward to catching up with our favorite mass spec scientists in a more relaxing setting!