At Emory, Size Selection Saves Time for NGS Prep
At the genome center at Emory University, scientists credit the Pippin Prep with shaving almost a full day off the sample prep process for Illumina’s mate-pair library prep.
Chad Haase, laboratory manager at the Emory GRA Genome Center, says that the Sage Science Pippin Prep size selection instrument his lab acquired about a year ago has replaced the 16-hour runs that had previously been done on gels with a very low agarose concentration.
Jamie Davis, a scientist at the genome center, says that she typically runs the Pippin “after we do our first end repair and the biotinylation reaction.” Size selection generally takes about an hour, she says.
The Pippin Prep also works well for 454 sequencing. Haase’s team had tried another automated gel system, but it wouldn’t work for fragments larger than 1 kb. “Once we started making the large mate-pair libraries — 3 kb, 8 kb, and 20 kb fragments — then we had to come up with something different and the Pippin system was the best,” he says.
“With the large kb mate-pair libraries, the Pippin has really been our savior there,” Haase says. “A sample prep process that used to take us three days, we can now get down to roughly two days.”
Haase says that his team also uses the Pippin before amplicon sequencing on the Roche/454 GS-FLX Titanium. “If you have any small fragments whatsoever in a 454 amplicon library, it’s going to ruin the run,” he says. “The emulsion PCR prior to sequencing is preferential to smaller fragment amplification, so you’d get a whole bunch of 50-base-pair sequencing reads” without using the Pippin for size selection.
New Cassettes Use Internal Standards
We’ve just released new dye-free cassettes for the BluePippin and Pippin Prep that use dye-labeled DNA markers* as internal standards, instead of typical external marker set. In the new product, the labeled markers will come premixed wi
th the Ficoll loading solution, and will be mixed and loaded with the sample DNA in each lane. The internal standards are designed to run well ahead of sample DNA targeted for elution, so that little or no marker contamination of eluted samples should occur.
These new cassettes will offer the following benefits:
- Faster run times. Internal standards automatically correct for the lane-to-lane mobility variation that occurs at higher voltage. Using the new high voltage 1.5% cassettes, collection of DNA fractions as large as 500 bp can be carried out in 35 minutes or less.
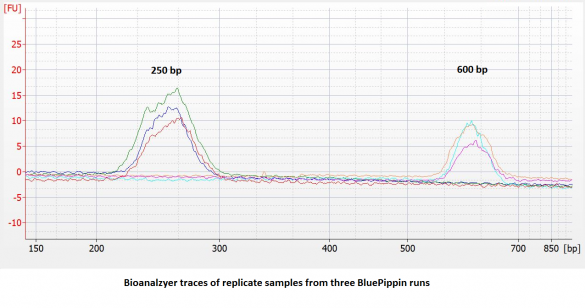
- Improved accuracy and reproducibility. Compared to dye-free cassettes run with an external marker, the accuracy and reproducibility of DNA extractions** will be improved (~ 5% vs. 10%). We have found the reproducibility to be quite good:
- 25% higher sample capacity. All five lanes can be used for samples. (This also means the per-sample cost goes down by 20%.)
- Use fewer lanes per run without penalty. Users may run a single sample, re-tape the open ports and save the remaining 4 lanes for later. With external standards, each run requires at least 2 lanes: one for external marker and one for sample.
Will internal standards contaminate your sequencing result?
The BluePippin internal standards are designed to run well ahead of the usable size range of the cassette. This minimizes the chance for marker contamination in eluted sample DNA since the platform excels at eliminating sample contamination from lower molecular weight fractions. In addition, when fractionating adaptered libraries, sequencing artifacts caused by marker DNA contamination are expected to be minimal, since the marker fragments are not complementary to amplification primers used by the major sequencing platforms. This expectation has been demonstrated on the Illumina platform (Tech Note in process). To enable identification/filtering of marker sequence, we’ve posted the sequences on our support page (www.sagescience.com/support). You can click the link below to get there:
http://sagescience.com/wp-content/uploads/2014/08/Internal-standard_sequences-UPDATED-082014.pdf
Input load and accuracy
The 1.5% DF cassettes can accommodate up to 10 ug of genomic input DNA. However, there are load-dependent changes in DNA mobility in these cassettes. Since most of our customers are running DNA samples of 2ug or less, we have changed our standard input load for calibration to 2 ug. Most users will find excellent accuracy for input loads up to 2 ug. At higher input loads, users should be prepared to run some preliminary tests to determine the best settings for their application. In general, at loads greater than 2 ug, selected DNA size will be slightly higher than programmed. As input load increases, so will the deviation from the programmed value.
Look or ask for these cassettes for the BluePippin:
For the BluePippin
- BDF1510 – 1.5% agarose with internal standards for the BluePippin. 250 bp-1.5 kb.
- BDF3010 – 3% agarose with internal standards for the BluePippin. 90 bp-300 bp.
For the Pippin Prep
- CDF1510 – 1.5% agarose with internal standards for the Pippin Prep. 250 bp-1.5 kb.
- CDF3010 – 3% agarose with internal standards for the Pippin Prep. 90 bp-300 bp.
* We use Mirus fluorescent label with our markers. “www.mirusbio.com”
** Accuracy = Deviation of the actual target base pair value from software input value divided by the actual value expressed as a percentage. (In calibration, actual DNA sizes are determined using an Agilent Bioanalyzer 2100.)
Reproducibility = 2X standard deviation of replicate samples expressed as a percentage of the average value.
“Overflow Detection” definition on the Pippin Prep to be eliminated
| |
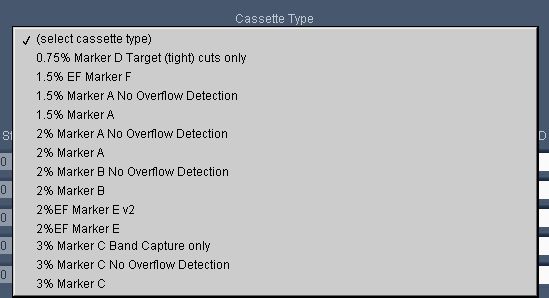
Pippin Prep users are likely familiar with our “overflow detection” cassette definitions that are found in the “Cassette Type” drop down menu of the Protocol Editor. We use these definitions because Pippin cassettes exhibit a phenomenon
where the elution module liquid volume increases with prolonged sample collections*. To prevent users from inadvertently overflowing the elution modules, we embedded limits in the software to restrict users from programming broad collection ranges.
However, overflowing is remedied by sealing the elution port with tape (which we now provide with our cassettes, or any PCR tape is fine). This keeps the volume constant and prevents the elution module from overflowing. For users using taped elution modules we removed the programming limits, and created the “No Overflow Detection” cassette definition. As the cassette list becomes more extensive, we’re trying to streamline options to make things easier and more uniform for users . Here’s a screenshot of our current list:
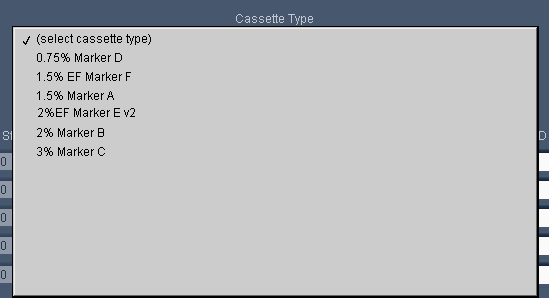
For our next software release, we’ll simplify the definitions (see below) such that they all effectively have “No Overflow Detection” – so please be careful. We recommend using tape for all extractions. If you don’t, be careful, or use the “Tight” programming mode.
*Overflowing is due to an electro-osmotic effect caused by the properties of the ultrafiltration membrane in the elution module.
ChIP-Seq sample prep tip – Interpreting the apparent presence of a secondary band in size-selected DNA
ChIP-Seq analysis is used to study the control of gene expression by identifying DNA regions that are associated with chromatin at the time of cellular analysis. The ChIP-Seq procedure typically involves cross-linking DNA to the associated chromatin proteins, sonication to fragment the chromatin, and immunoprecipitation of chromatin using an antibody to DNA Polymerase II or a transcription factor. The DNA regions associated with chromatin are identified by DNA sequence analysis. To prepare the DNA for sequencing one must dissociate and purify the DNA away from chromatin, create blunt ends, add poly(A) tails, and ligate adaptors for PCR amplification.
Importance of size selection with high yield
A DNA size selection step is done to improve the quality of the input sample for sequencing. PCR amplification is performed for a limited number of cycles (18-20) to increase the amount of material available for sequence analysis. Using the Pippin Prep for DNA size selection, one can get a tight size distribution and a high yield (50-80%) in the selected size range. The high yield is particularly important because it means that fewer PCR cycles are needed, which in turn means that the amplified sample will have less sequence bias and be more representative of the original DNA sample.
Appearance of an apparent ‘secondary’ larger band after DNA size selection
Because of the very small amounts of DNA present in ChIP-Seq samples, amplification is sometimes performed before rather than after the size selection step. In these cases some users have been surprised to find a secondary band of larger size when the size-selected and PCR-amplified DNA is analyzed on an e-Gel (Life Technologies) or FlashGel (Lonza) system. What could be leading to this observation?
One plausible explanation for this phenomenon is the presence of some single-stranded DNA (ssDNA) in the amplified DNA. A certain amount of ssDNA can be generated among the dsDNA reaction products of PCR if too many cycles of amplification are performed or if the primer concentration is too low (1). Below we present our interpretation of this phenomenon. We also welcome you to comment on this blog post and share your personal experiences that may be pertinent.
How can over-amplification lead to ssDNA in the sample?
A ChIP-Seq library contains a tiny amount of immunoprecipitated DNA, so it is tempting to over-amplify to be sure of getting sufficient material for sequence analysis. Unlike traditional PCR in which a single-copy gene is amplified from a small amount of DNA, all of the ChIP-Seq DNA contains adaptors and is eligible for amplification using the universal primers. Thus the primers are quickly consumed. As the primer concentration becomes limiting, one of the two PCR primers may be better able to anneal than the other, and this can lead to accumulation of some ssDNA within the population of amplified dsDNA (2).
Furthermore, in the later cycles the amplification products can accumulate as denatured ssDNA strands. (The two strands can fail to reanneal because the amplicons are diverse in insert sequence even though they have the same primer binding sites. Therefore most or all of the library amplicons may not be present at sufficient concentration to reanneal during the lower temperature steps in the PCR cycle.
In a preparative agarose gel system (with a low voltage gradient) ssDNA fragments could potentially co-migrate with dsDNA fragments. When a portion of the size-selected sample is then run on an analytical agarose gel (with a high voltage gradient) ssDNA fragments could potentially migrate differently than dsDNA fragments. Personal communications from some users tell us that this explanation is plausible. When a ‘secondary’ DNA band of apparently larger size was re-amplifiied and analyzed on an analytical gel, only the smaller, primary band was generated unless the sample was over-amplified. In this case, sequence analysis also verified that the sequence of the upper and lower bands were indistinguishable.
Summary recommendations
In general, limit the number of PCR cycles to 18-20. If ‘secondary’ DNA bands are seen in the size selected sample, try reducing the number of PCR cycles. This may have the added benefit of reducing PCR-induced sequence bias.
Contribute your experiences! As mentioned above, we encourage you to comment on this blog post and share any experiences that may be pertinent to the discussion.
References
- Roux, K.H. Optimization and troubleshooting in PCR. Genome Res. 1995 4: S185-S194 Link
- PCR Troubleshooting web site published by The PCRCore Facility at Children’s Hospital of Philadelphia Link
![]() Follow @DNAsizing on Twitter
Follow @DNAsizing on Twitter