AGBT 2019 Re-Cap
AGBT 2019 was a smashing success this year. Featuring a triumphant return to Marco Island FL with postcard perfect weather, the science was (as usual) top-notch. The renovated Marriott greatly upgraded its conference facilities and added an attractive new tower with roof top pool and game room.
By way of recap, single-cell sequencing continues to be a dominant topic. We particularly enjoyed the talk by Jiannis Ragoussis from McGill University, providing a comprehensive expression analysis of 20,000 Glioblastoma cells, offering insight on how significant these tools can be for medical research. Spatial profiling created the biggest buzz, and the Broad Institute’s Evan Macosko’s presentation on Slide-Seq (a method where RNA is transferred from freshly frozen tissue sections onto a surface covered in DNA-barcoded beads) generated a lot of interest.
Surprising to us – if that concept can even be applied to AGBT – were the advances in genomic structure and straight up genome sequencing. We saw quite a bit of Hi-C data: Wendy Bickmore from the University of Edinburgh gave a great talk (“spatial organization of the human genome”) looking at long-range gene regulation, and Katherine Pollard from the Gladstone Institute at UCSF (“A population view of human chromatin structure”) gave a biophysical approach to examining the effect of mutations on structure. Ting Wu from Harvard provided a fascinating update (“looking at chromosomes”) on direct visualization of chromosome structure using Oligopaints and other novel methods.
For excellence in genome sequencing, NHGRI’s Adamy Phillipy’s talk, “Telomere-to-telomere assembly of a complete human X chromosome” was inspiring and used many long-range and long-read techniques, including the Oxford Nanopore Promethion (in collaboration with UC Santa Cruz’ Karen Miga, who gave a separate talk on their pipeline). Mike Hunkapiller from PacBio showed very nice gapless assemblies of the notoriously difficult SMN1 and SMN2 genes using the closed consensus sequencing method on the Sequel platform.
As for us at Sage Science, we had a relaxing beach-side Lanai suite. Aside from co-hosting a Queen-the-band themed party with Seqwell (Genomian Rhapsody), we presented a SageHLS poster that used the HLS-CATCH method to sequence the PKD1 pseudogene (our other in-suite posters can be viewed here). We believe that HLS-CATCH purification of long genomic targets can be a great tool for helping resolve difficult regions in the genome – like the aforementioned pseudogenes, other repeat elements, and SVs– and hopefully helping efforts to produce more complete genomes and understanding the function of genome structure.
Obviously, it is impossible to truly summarize such an intensive meeting- but we would like to give an additional shout out to John Charles from NASA who came down to give an update on Mars Mission plans– what could be cooler than that? On another note, next year’s meeting will mark the 20-year anniversary of AGBT. We’re sure there will be something extra-special in store, look forward to more great science!
Linked-Read Sequencing Advances Understanding of Cancers
A new study* from the University of Connecticut Medical School, Jackson Labs, and collaborators demonstrate the utility of using emulsion based linked-read sequencing (10X Genomics) for cancer research. Published in January’s Otology & Neurotology studies patients with Neurofibromatosis Type 2 (NF2) – a disease that manifests as benign brain tumors in the sheath of the cranial nerve VIII, typically causing hearing loss. The researchers compared DNA from five patients with fast-growing tumors with DNA from five patients slow-growing tumors and DNA from matching blood samples.
Using whole genome linked-read sequencing, results identified several large deletions (ranging from 5 to 650kb) in the NF2 locus correlating to the severity of the disease phenotype. The study reveals other correlating structural variants in a number of genes including FBXW7 (implicated in tumorigenesis in many other cancers) and TSPAN (implicated in esophogeal cancer). Interestingly, 4 of 5 of the high-growth tumor patients showed deletion in the VEGF-C locus. Citing a number of studies and trials of the anti-VEGF drug bevacizumab, which targets VEGF-A, the authors believe that the VEGC-C result supports previous findings that suggest that it could be predictive of treatment response.
From a methods standpoint, linked-read sequencing requires only 1 ng of DNA input and produces haplotype phasing information. For this study, our PippinHT was used to filter away smaller DNA fragments (using the >40kb High Pass protocol) to maximize the efficiency of the linked-reads for long range SV analysis. With a 1 ng input requirement, there is ample recovery in the PippinHT – users simply quantify the sample with a Qubit fluorometric assay, and dilute the sample accordingly. Concentration or buffer exchange is not necessary. On a side note, our SageHLS platform has the capability to provide very large targeted genomic regions, a great application for linked-reads given the low input requirement (read a preprint about this here).
*Linked-read Sequencing Analysis Reveals Tumor-specific Genome Variation Landscapes in Neurofibromatiosis Type 2 (NF2) Patients
Roberts, Daniel S. et al., Ontology & Neurotology: February 2019 – Volume 40 – Issue 2 – p e150-e159
doi: 10.1097/MAO.0000000000002096
SageELF Size Selection for PacBio Circular Consensus Sequencing
A new preprint has landed on BioRxiv that reports on high-accuracy circular consensus sequencing (CCS) on the PacBio Sequel. The study, “Highly-accurate long-read sequencing improves variant detection and assembly of a human genome”, is authored by PacBio and an impressive team of collaborators featuring notable bioinformaticians and members of the Genome in a Bottle Consortium. The data suggest that with CCS, very accurate (Q30) DNA sequence can be obtained from a single >10kb molecule (read PacBio’s blog on the study here)
The gist of the method is this: processivity improvements have yielded polymerase read lengths of approximately 150kb. Since SMRTbells are circular, a polymerase should be able pass a 15kb DNA fragment 10 times, and re-reading the molecule 10 times should yield a 99.9% accurate sequence.
 Our customers may be aware of our High-Pass library size selection with the BluePippin in which, average read lengths can be improved – often doubling N50s. But for CCS, its crucial to prepare libraries with relatively uniform size, with the goal of producing a full run of Q30 15kb reads to be assembled against a reference. For this, the SageELF DNA fractionator is the tool for the task. The SageELF produces narrow fragment size distribution, reproducibly, and provides a great deal of flexibility. For instance, users can run a 15kb library and archive a 20kb library from another well. Or, adjacent wells can be pooled, increasing library amount with only a slight widening of the distribution range.
Our customers may be aware of our High-Pass library size selection with the BluePippin in which, average read lengths can be improved – often doubling N50s. But for CCS, its crucial to prepare libraries with relatively uniform size, with the goal of producing a full run of Q30 15kb reads to be assembled against a reference. For this, the SageELF DNA fractionator is the tool for the task. The SageELF produces narrow fragment size distribution, reproducibly, and provides a great deal of flexibility. For instance, users can run a 15kb library and archive a 20kb library from another well. Or, adjacent wells can be pooled, increasing library amount with only a slight widening of the distribution range.
In BioRXiv paper, the follow protocol was used:
1. Start with 3-4 ug DNA
2. Shear the DNA with Diagenode Megarupter to 15-20kb
3. Construct SMRTbell libraries
4. Size Fractionate with the SageELF, collect a 15kb fraction
5. Run on the Sequel
The following fractionation protocol was used (this is not detailed in the publication, but based on private communication with the authors):
1. Use cassette kit #ELD7510 (for 1-18 kb fractionation)
2. Load 1-2 ug/run (fractions were pooled from two runs)
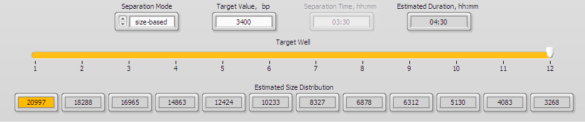
3. Enter 3400 into well 12 in the protocol editor (below)
4. A 15 kb fraction will be found in well 4, approx. 4.5 hour run
We do have alternative recommendations:
1. We offer cassette kit #ELD4010 (for 10-40kb fractionation). This should provide an even narrower size distribution than the 0.75% agarose cassettes.
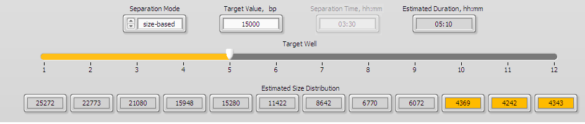
2. Enter 15000 into well 5 in the protocol editor. The 15 kb fraction will be found in well 5 (and a 20kb fraction will be found in well 3).
3. 3-4ug of input DNA in one lane should be sufficient so pooling from two runs may not be needed.
Product Update: The High Pass Plus Cassette
The High Pass Plus™ gel cassette is the newest addition to the Pippin Family. As the name suggests, it is dedicated to our BluePippin “High-Pass” DNA size selection which has been a go-to method for increasing the read lengths for long-read sequencing.
High Pass size selection removes smaller DNA fragments from a sheared genomic DNA (or sequencing library) while collecting the remaining larger fragments above a tightly controlled size threshold. This way, larger molecules can be presented to the detector or droplet, and better sequencing performance can be achieved.
Long-read sequencing sample prep has been improving overall, in terms of set-up time and workflow. We decided to look at the High Pass as well to see if we could optimize the approach. To this end, we designed an entirely new gel cassette dedicated to High Pass– the High Pass Plus. We’re happy to say that we were able to cut the runtime in half and improve performance and yield.
Here’s a comparison between the standard BluePippin Cassette and the High Pass Plus:
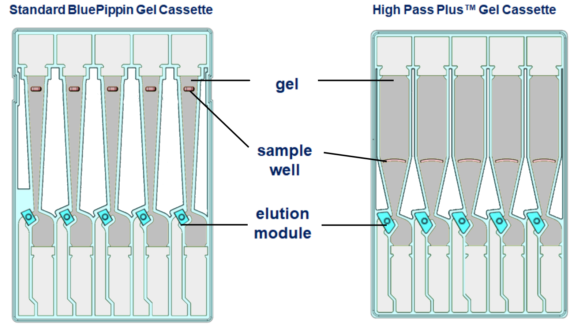
The High Pass Plus cassette has a stocky separation column, so DNA has a shorter distance to travel. The wider column and increased taper provide higher resolution for a cleaner and more accurate size cut-off. The larger sample wells now allow a maximum load of 10ug (100% up from standard Blue Pippin cassettes). We’ve also increased the size of the elution module and surface area of the filtration membrane, bringing about improved sample recovery and reproducibility.
Here’s what we were able to accomplish, when compared to the current BluePippin standard:
• Half the run time
• Twice the loading capacity
• Better recovery and reproducibility
We offer a >15kb High Pass Plus at this time. Next up will be a >20kb and >30kb.
Blue Pippin software requirement is v6.31/6.40 CD31. Available here:
The High Pass Plus cassette is available now, order number BPLUS10, or if you’d to purchase a 3 pack (BPLUS03) to try let us know.
Tracking Yellow Fever: New Study Aims To Tackle YFV Epidemics
Last year, we posted a story about an international team of scientists who embarked on a mission with Brazilian researchers to study the dangerous mosquito-borne Chikungunya virus (“Tracking Chikungunya: New Study Traces Outbreak Path”- we were pleased to donate a Pippin Prep to the effort.)
Recently the team published a new study in Science, having deployed NGS to elucidate the epidemiology of the Yellow Fever virus in Brazil: “Genomic and epidemiological monitoring of yellow fever virus transmission potential”. According to the authors, virological surveillance requirements are to “ (i) track epidemic origins and transmission hotspots, (ii) characterize genetic diversity to aid molecular diagnostics, (iii) detect viral mutations associated with disease severity, and (iv) exclude the possibility that human cases are caused by vaccine reversion.” By sampling humans and non-human primates (NHP) across the state of Minas Gerais, the epicenter of a 2017 outbreak, the researchers analyzed how the virus spreads through space, between humans and NHPs, and the “contribution of the urban cycle”. The authors note that this type of real-time monitoring can contribute to global efforts to eliminate future epidemics – and one would assume, potentially save lives.
Our minor contribution to this work was based on outreach from Antonio Charlys Da-Costa, who had used a Pippin Prep as part of his studies at Dr. Eric Delwart’s lab at the UCSF Blood Systems Research Institute. Dr. Da-Costa, now at the Sao Paulo Institute of Tropical Medicine, was able to marshal resources from a number of other suppliers for this cause, including Illumina, Zymo Research, and Promega (surely there were others, apologies for the non-mention). Congratulations to the whole team, it’s inspiring to see such diverse institutions and agencies pulling these large-scale efforts together – and individuals like Antonio who went the extra mile.
An interesting side note, a number of other important viral genomic findings were also published during this time by a consortium of Brazilan labs in collaboration with Dr. Delwart and UCSF:
Wuhan large pig roundworm virus identified in human feces in Brazil