AGBT23 (Broad Institute):
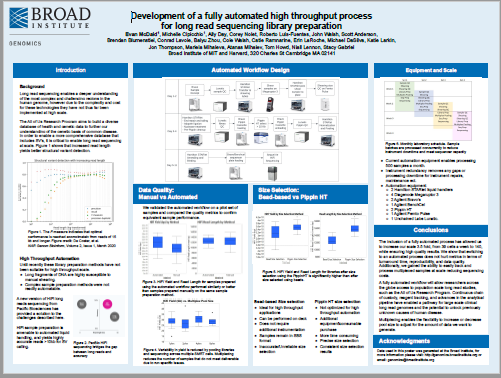
Development of a fully automated high throughput process for long read sequencing library preparation
From Broad Institute, this poster describes their fully automated pipeline for producing PacBio Hi-Fi Whole Genome Sequences for the NIH’s All of US program. PippinHT is used for library size selection
AGBT23 (Stanford):
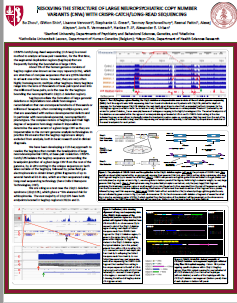
Resolving the structure of large neuropsychiatric copy number
variants (CNVs) with CRISPR-CATCH/long-read sequencing
This update of a collaboration betwee Stanford, Katholieke Universiteit Leuven, Belgium, and the Mayo Clinic uses CATCH to look at Segmental Duplications that flank the the neuropsychiatric 22q11.2 deletion region

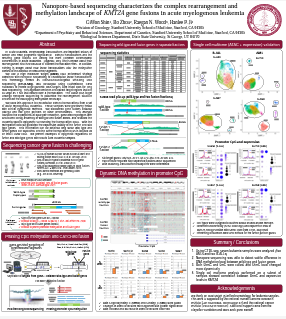
AGBT23 (Stanford University):
Nanopore-based sequencing characterizes the complex rearrangement and methylation landscape of KMT2Agene fusions in acute myelogenousleukemia
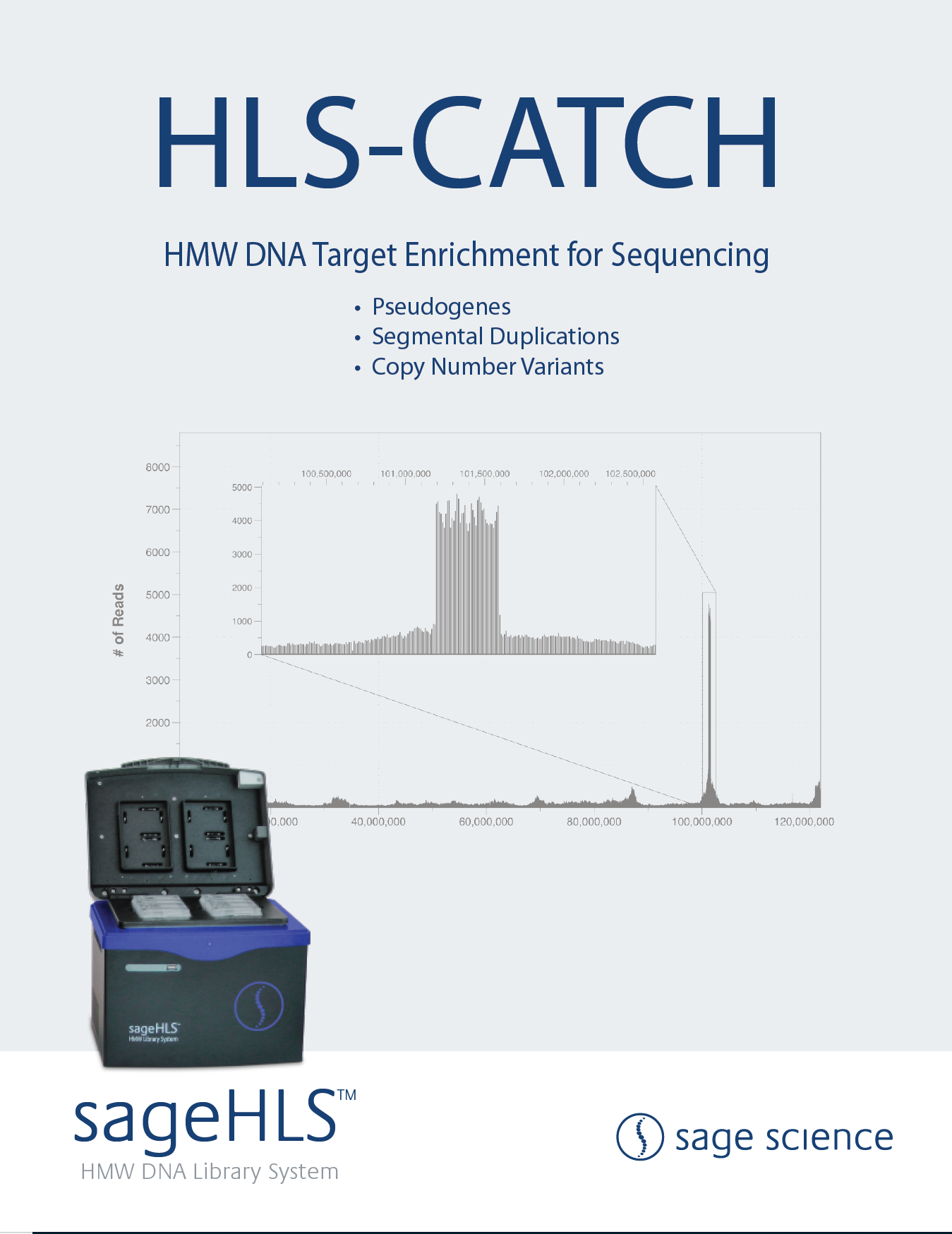
This poster employs HLS-CATCH with nanopore sequencing to evaluate translocations in the KMT2A gene associated with acute myelogenous leukemia. Nanopore sequencing provides valuable epigenetic methylation patterns.
AGBT23 (University College London):
Cas9 targeted long-read nanopore sequencing of ABCA4 and the OPN1LW/OPN1MW gene array for investigation of inherited retinal disease
From the Institute of Ophthalmology, University College London, this poster describes using the HLS-CATCH process to evaluate complex rearrangements in genes associated with retinal disease.

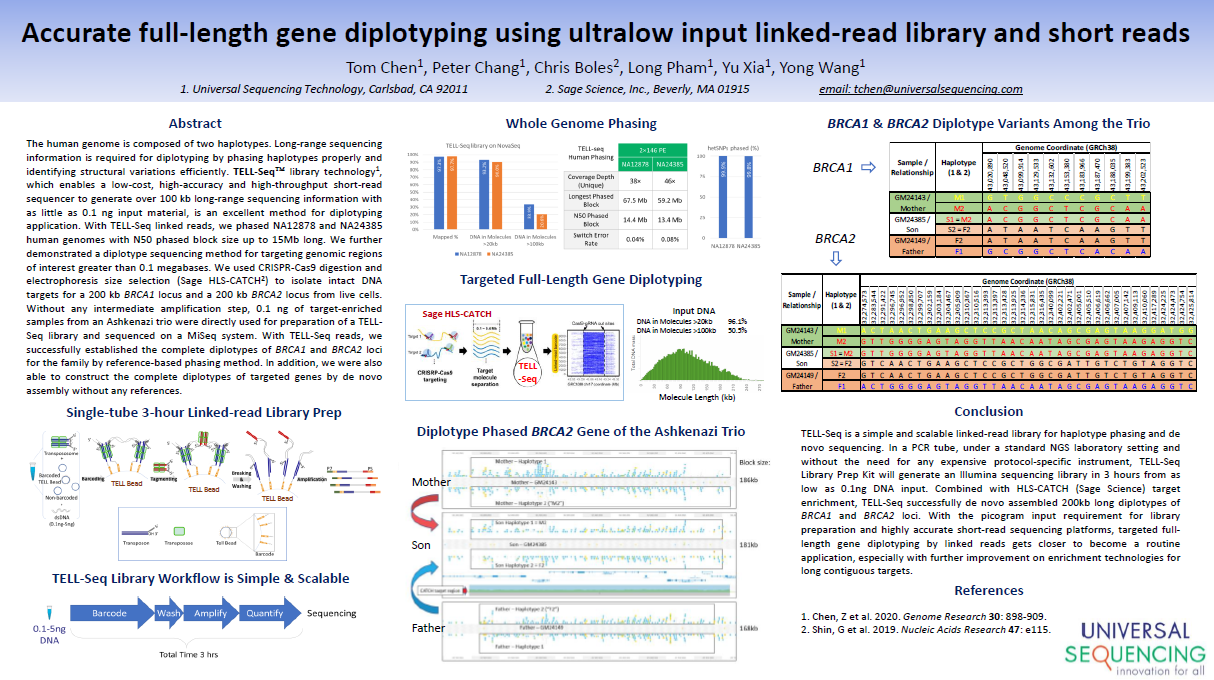
AGBT23 (Universal Sequencing):
An Optimized Single-Tube Linked-Read Method Empowers Short-Read Sequencers to Phase 2 to 200 Kilobase DNA targets
The TELL-Seq linked read process is demonstrated. High quality phasing information provided for 200kb targets enriched with HLS-CATCH (HLS2 system), and for smaller targets enriched with other methods (Exo/pulldown and PCR).

(Earlham Institute):
High-Pass DNA Size Selection for Recovery of a Highly Degraded Sample for Oxford Nanopore Long-Read Sequencing
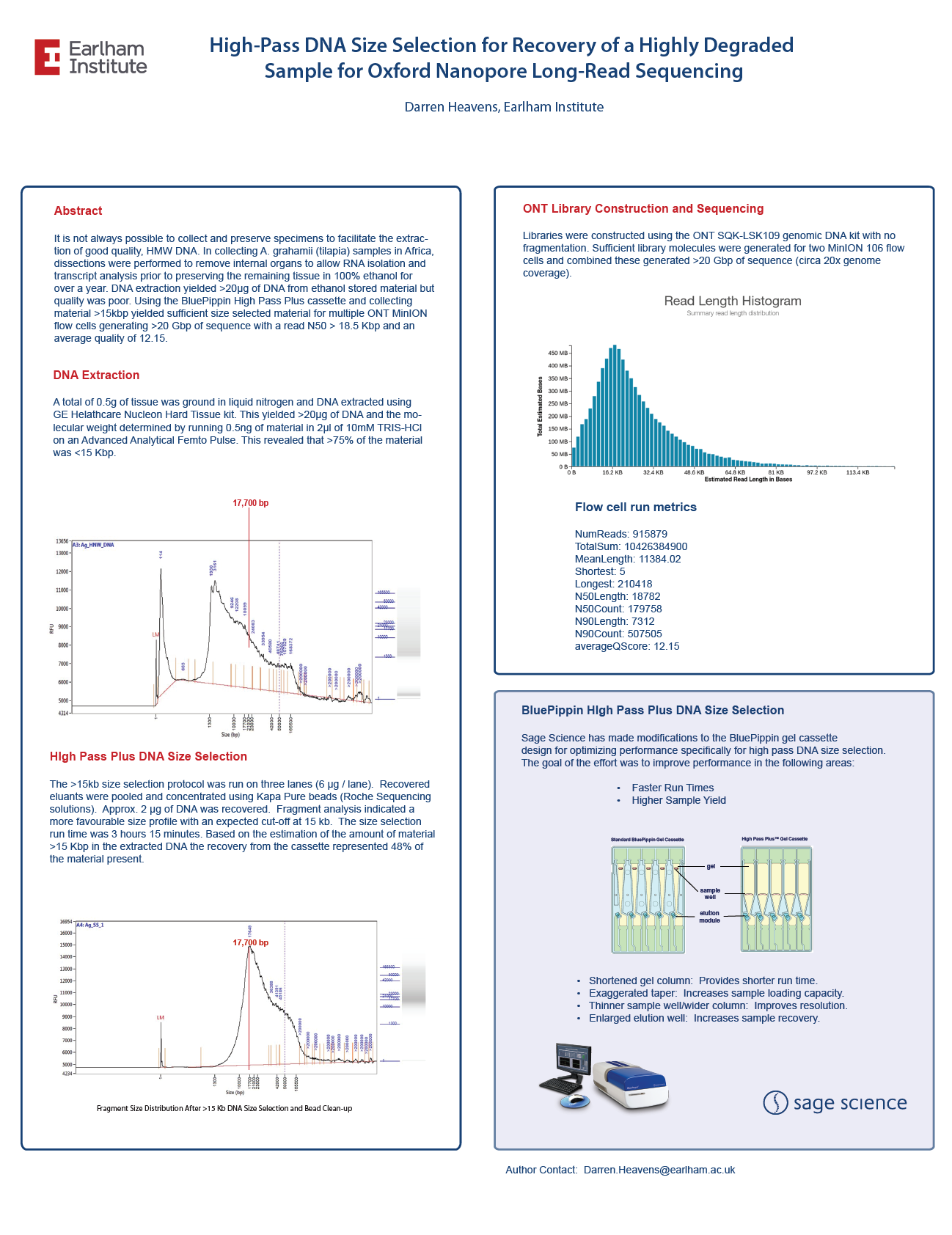
From the Earlham Institute, a highly degraded fish DNA sample was size selected with the BluePippin High Pass Plus cassette. Recovered DNA was sequenced on a Oxford Nanopore Minion to 20X genomic coverage.

(Sage Science):
Sage Products Powering New Methods, 2018-19
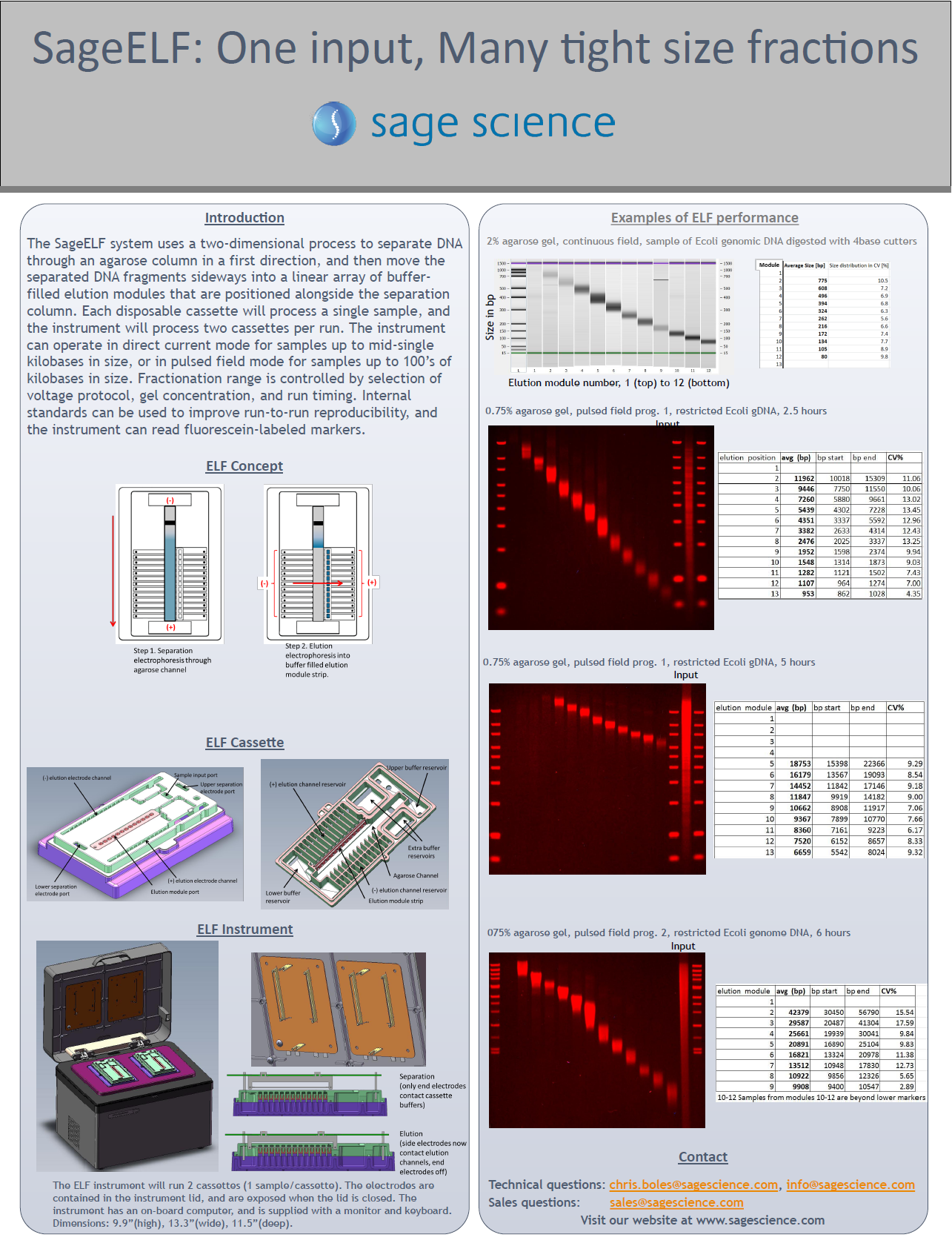
Summarizes recent applications of Sage Science Instruments. The SageELF size fractionation is used to tightly sized libraries for PacBio Sequel closed consensus sequencing (CSS). The PippinHT is used for size selection of circulating tumor DNA from plasma.