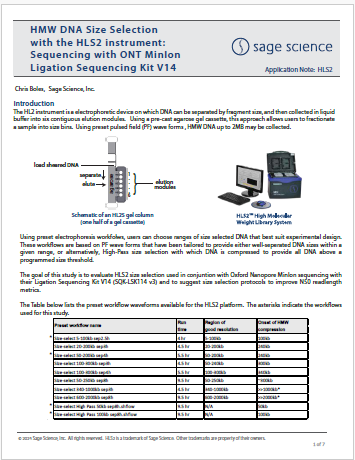
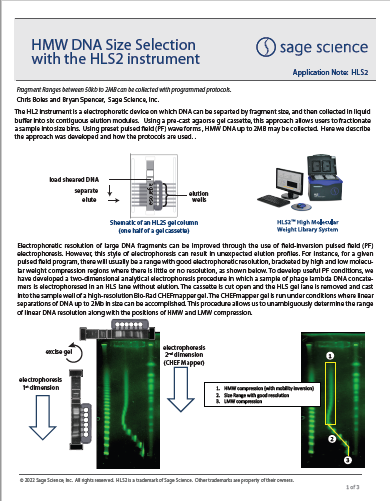
HMW DNA Size Selection with the HLS2 instrument: Sequencing with ONT MinIon Ligation Sequencing Kit V14
This study evaluates HLS2 size selection used in conjunction with Oxford Nanopore MinIon sequencing with their Ligation Sequencing Kit V14 (SQK-LSK114 v3). Size selection protocols to improve N50 readlength
metrics are suggested.

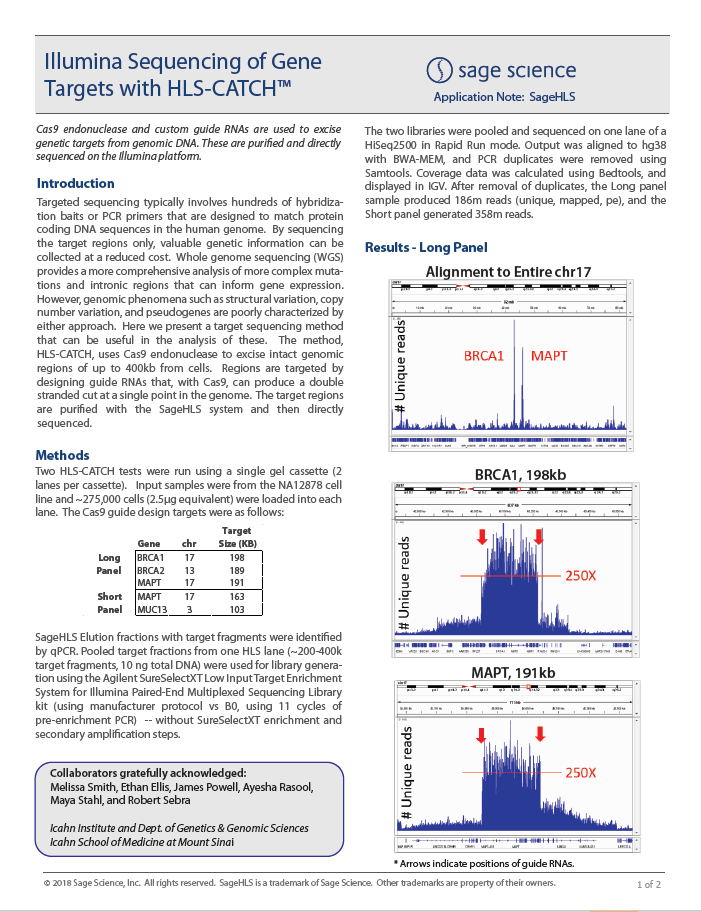
ASHG ’23: Detection of large inversions involving the Lynch syndrome gene PMS2 using Cas9-assisted targeted sequencing.
In a collaboration between Sage and Illumina, Sage’s HLS-CATCH process was used with Illumina’s Complete Long Read (CLR) process to identify a large inversion on the PMS2 locus in the Ashkenazi trio. Using Coriell cell lines HG002, HG003 and HG004, it was shown that the maternal haplotype inherited by the son carries this inversion.
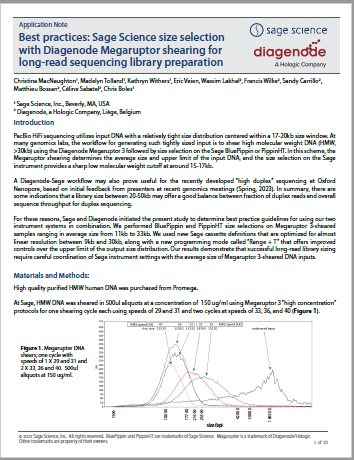
Best practices: Sage Science size selection with Diagenode Megaruptor shearing for long-read sequencing library preparation
This is an in-depth collaboration between Sage and Diagenode/Hologic. Megartupter 3 DNA shears of different sizes were subject to size selection as part of our “Range+T” evaluation for PippinHT and BluePippin. Data is provided which provide the best practices for achieving DNA size selections with tight fragment distributions around median sizes from 9-30kb for long-read sequencing applications.
Range + T Size Selection Method
Increases PacBio HiFi Read Lengths
From the Arizona Genomics Institute, University of Arizona College of Arizona College of Agricultural & Life Sciences. Researchers were given early access to the Range + T programming mode. The PippinHT was used for size selection on PacBio SMRTbell library construction for HiFi Sequencing of plant genomes. High Quality sequencing reads were obtained with mean subreads of 20-25kb, indicating that Range + T can increase current HiFI readlength recommendations.

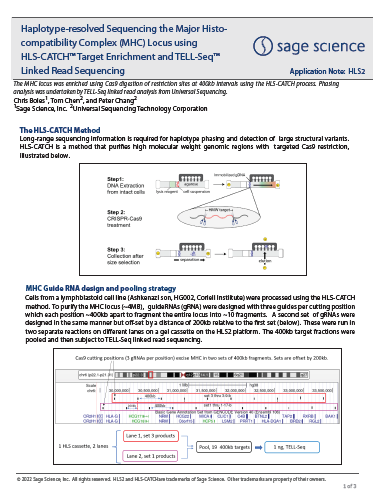
Haplotype-resolved Sequencing the Major Histocompatibility Complex (MHC) Locus using HLS-CATCH™ Target Enrichment and TELL-Seq™ Linked Read Sequencing
From Universal Sequencing and Sage Science, HLS-CATCH and UST TELL-Seq linked reads are used to Haplotype-resolve the entire MHC complex. The MHC locus was purified as 20X 400kb fragments using the CATCH process.


Accurate Cost-effective Haplotype-resolved Sequencing of Large Targeted Genomic Regions Using HLS-CATCH™ Sample Prep with TELL-Seq™ Library Preparation
From Universal Sequencing and Sage Science, HLS-CATCH and UST TELL-Seq linked reads are used to Diplotype full-length BRCA2 targets from a model Ashkenazi Trio
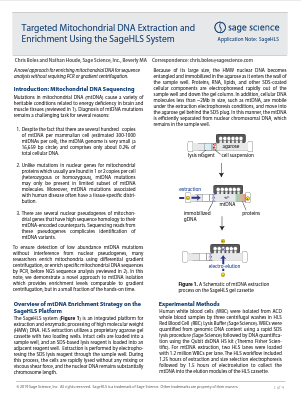
PippinHT: Collection of <167 bp DNA for Cell-Free DNA Library Construction
Parameters for timed DNA size selection for collection of fragments below 167bp are provided for the PippinHT platform. Studies have shown that shorter cfDNA fragments (40 -167bp) retain genetic information about the fetus,
tumor associated copy number aberrations or the cellular origin of cfDNA,