Request a quote or order online.
More questions?
Contact Us.
The Kit
Q: On which Illumina platforms can TELL-Seq libraries run?
A: All Illumina platforms except the iSeq.
Q: What is the cost of TELL-Seq library prep for human WGS?
A: <$180
Q: How do you order the Kit?
A: Kits can be ordered on the Sage Science online store. The basic kit has two boxes; one with TELL-bead barcodes and the other contains the barcoding enzyme mix. Users also need to purchase a Sequencing Primer kit (for Illumina) and a Library Multiplex Primer Kit. The basic barcode kit can be used for 4-12 genomes, depending on size. The two Primer kit types can be used for multiple libraries beyond those in the basic kit, depending on the genome types that are sequenced.
Q: In what ways does TELL-Seq differ from 10X Genomics linked-reads?
A: TELL-seq uses a transposase to insert barcodes into double-stranded DNA, 10X linked-reads partition single-stranded DNA to which barcodes are ligated. TELL-Seq has a much higher barcode capacity (>2 billion vs 4 million) and can accommodate smaller (<200Mb) genomes. TELL-seq does not require non-standard lab instrumentation or supplies and costs roughly half of the library construction cost (for human WGS).
DNA
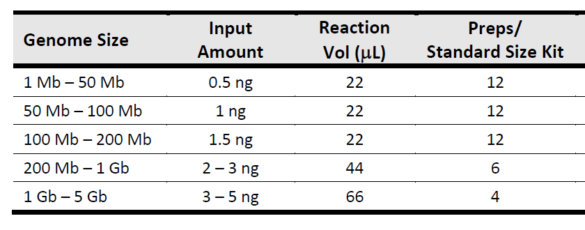
Q: What is the DNA input requirement?
A: 0.5 ng – 5 ng. 5ng is recommended for human WGS.
Q: How should I prepare my DNA?
A: HMW weight DNA extraction kits (ie. Qiagen Magattract) are best. Larger DNA fragments will provide larger phase blocks.
Q: Does TELL-Seq work with HLS-CATCH?
A: It is a great option because of the low DNA input requirement. We were able to sequence the BRCA2 gene, on a 200kb target, from a trio using the small genome-scale. Here’s the whitepaper.
Workflow and Analysis
Q: How is TELL-Seq linked read sequencing analyzed?
A: Universal Sequencing Technology (UST), the developers of TELL-Seq, have a software pipeline to prepare linked reads. These can be analyzed by UST phasing or assembly pipelines, or converted to a 10X Genomics format and analyzed by Longranger or Supernova and visualized with the Loupe browser.
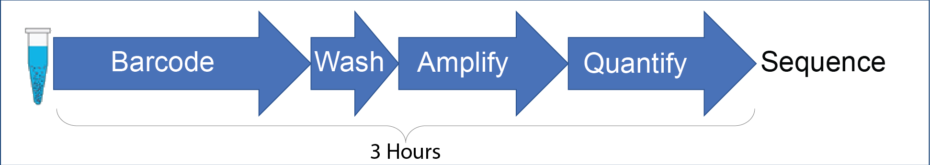
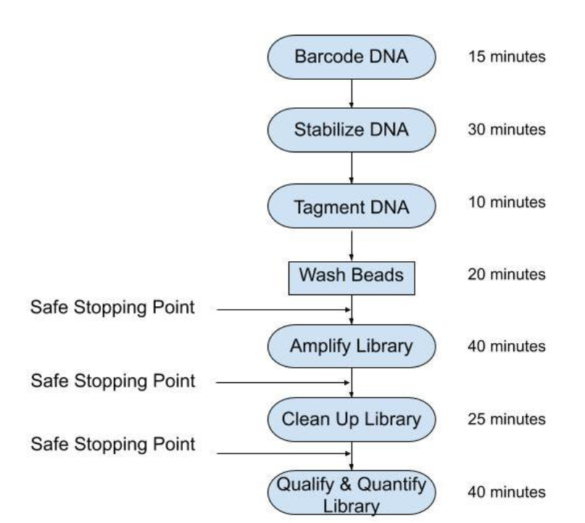
Q: Do I need to make a separate Illumina library from the barcoded DNA?
A: No, the Illumina library is ready to run after a 3-hour prep. The workflow is not significantly different than a typical Illumina library prep. It requires a similar amount of hands-on effort and the same standard lab instrumentation.