The Long…
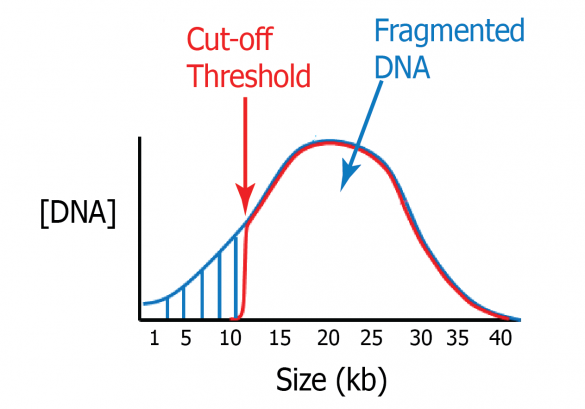
Size selection is perhaps even more important for long-read technologies than for short-read sequencers. Removing shorter fragments during library construction allows these sequencers to focus on the longest fragments, maximizing the read lengths generated during analysis.
To facilitate this, we developed the High-Pass protocol. Users select a cut-off threshold value, in base-pairs, and the Pippin system will collect all fragment sizes from the input sample that are larger than that value.
High-Pass protocols are available for the BluePippin and PippinHT systems.
BluePippin PippinHT
(4 samples/run, 5 ug max. input) (up to 22 samples/run, 1.5 ug max. input)
If you are using or considering PacBio, 10X Genomics, or Oxford Nanopore platforms, contact us about Pippin High-Pass size selection!
…And The Short Of It.
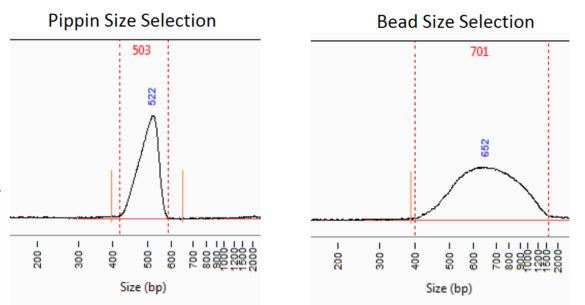
Automated size selection provides hassle-free alternative to bead-based or manual size selection or manual gel purification for Illumina workflows. It also results in a more accurately sized library, a narrower size selection if needed, and a tunable step to help optimize your method.
Short-read library size selection can be accomplished on every Pippin model:

Pippin Prep and BluePippin PippinHT
(5 samples/run, 5 ug max. input) (up to 24 samples/run, 1.5 ug max. input)