Update: Access the BioRxiv preprint of this study here..
With customized Cas9 nucleases, scientists can now isolate specific large genomic DNA fragments with the SageHLS platform for targeted library preparation in the 10x Genomics Chromium system.
This approach, known as HLS-CATCH, allows users to apply long-range genomics to their gene or genomic region of interest, characterizing and phasing not only single nucleotide variants but also large structural variants including deletions, inversions, and translocations.
The Sage Science team collaborated with Hanlee Ji’s lab at Stanford University to develop and validate this protocol. The data and figures shown below were provided courtesy of GiWon Shin at Stanford University. Check out GiWon’s presentation of this work from AGBT 2018.
Here’s how it works:
-
- Intact cells or nuclei are loaded into SageHLS cassettes
- Electrophoretic extraction leaves chromosomal-length genomic DNA immobilized in the sample well wall
- Custom Cas9 cleavases perform enzymatic DNA processing
- Size selection electrophoresis moves Cas9-cleaved target regions into the gel channel, away from uncut untargeted genomic DNA in the sample well
- Products are localized by qPCR after electroelution from the gel channel
- Captured DNA can be used directly in the 10X Chromium system for library construction
View or download the HLS-CATCH workflow schematic
The HLS-CATCH+10x Genomics workflow is cost-effective. For instance, a 200Kb target can be isolated using HLS-CATCH with as few as two custom Cas9 complexes, whereas hybridization-capture methods would require hundreds to thousands of probes. In addition, because of the complementarity between the HLS-CATCH method and the 10x Chromium library system, sequencing reagent costs to achieve 100x phased target coverage are more than 10-fold lower than that required to obtain 100x phased whole genome data.
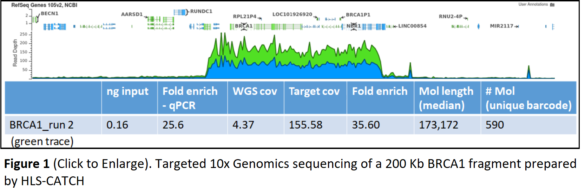
200 Kb Target: BRCA1
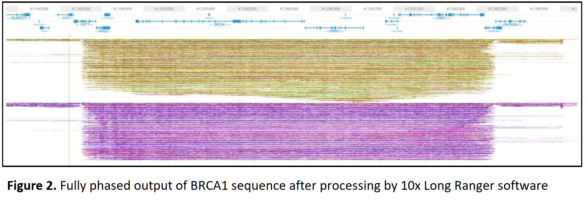
The study targeted a 200 Kb region including the human BRCA1 gene. With input material of 1.5 million diploid cultured cells, scientists recovered up to 50,000 copies of the BRCA1 fragment — at enrichments of 25-35-fold compared to a control non-targeted gene. Libraries were prepared on the 10x Genomics Chromium system and sequenced on an Illumina NextSeq500 system. The run produced 155x coverage of the targeted BRCA1 region, with only 4.4X coverage of the non-targeted remainder of the genome. Analysis with the 10x Long Ranger alignment software demonstrated that fully phased haplotypes could be determined.
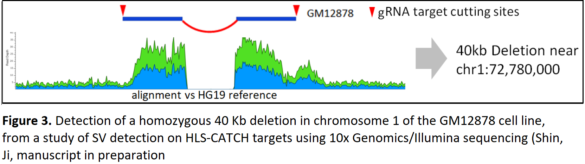
Structural Variants- GM12878 cell line
In another study, the team targeted large structural variants in the cell line GM12878. They isolated 40 regions, each 100 Kb long, in a single multiplex HLS-CATCH procedure.
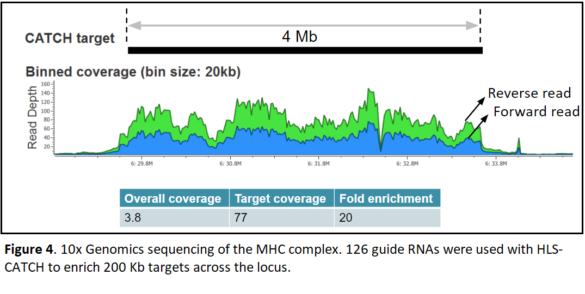
4Mb Target: MHC Locus
In a third study, the team used a set of 126 guide RNAs to tile across the entire 4 MB MHC region. Each Cas9 target was approximately 200Kb in length.
Summary
The new HLS-CATCH + 10x Genomics workflow offers significant new advantages for genetic research and testing. The method allows detection of both SNPs and large structural variants, in contrast to conventional hybridization-capture methods. 10x Genomics linked-read technology allows high coverage and fully phased variant maps over the HLS-CATCH targets.
We believe this study provides the first demonstration of a targeted sequencing workflow using genomic fragments that are greater than 20 Kb in length. The HLS-CATCH + 10x Genomics method also produced the first fully haplotype-resolved sequence of the entire 4Mb MHC region, a capability which opens the door to many new research areas in immunogenetics.