More applications are becoming possible thanks to the pairing of next-gen sequencing and cell-free DNA (cfDNA). Whether it’s circulating tumor DNA that helps scientists monitor tumor progression and treatment response or cell-free fetal DNA that makes it possible to perform noninvasive prenatal testing, the possibilities are fascinating.
A major challenge with cfDNA analysis, however, is isolating it from other small DNA fragments (for example, tumor from healthy cell-free DNA, or fetal from maternal DNA). Accurate, automated size selection with our Pippin family of instruments addresses this issue by allowing scientists to target just the size range associated with the cfDNA of interest.
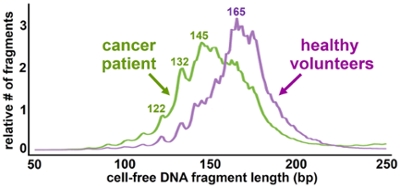
(Underhill et. al. PLOS Genetics, 2016)
In a bioRxiv preprint, scientists from Cancer Research UK and other institutions reported that adding a simple sizing step enabled important insight about tumor genetics in a cfDNA study. They used the PippinHT to target fragments 90-150 bp long, successfully separating them from the typical healthy cell-free DNA that peaks around 167 bp. Incorporating size selection provided as much as 11-fold enrichment for tumor DNA compared to a conventional pipeline, and allowed the team to spot clinically relevant copy number changes that could not be observed without size selection.
Separately, in work Sage Science conducted with Rubicon Genomics and reported in this poster, we used Pippin Prep to separate fragments larger than 160 bp from shorter fragments. Libraries were then prepared for sequencing using Rubicon’s ThruPLEX®Plasma-Seq kit, which is designed for low-input samples from plasma and liquid biopsies like cfDNA. Even though we started with less than 1 ng of size-selected material, we were able to produce highly diverse libraries that were significantly enriched for cell-free DNA fragments between 60 bp and 120 bp. That enrichment did not happen for libraries prepared without size selection.